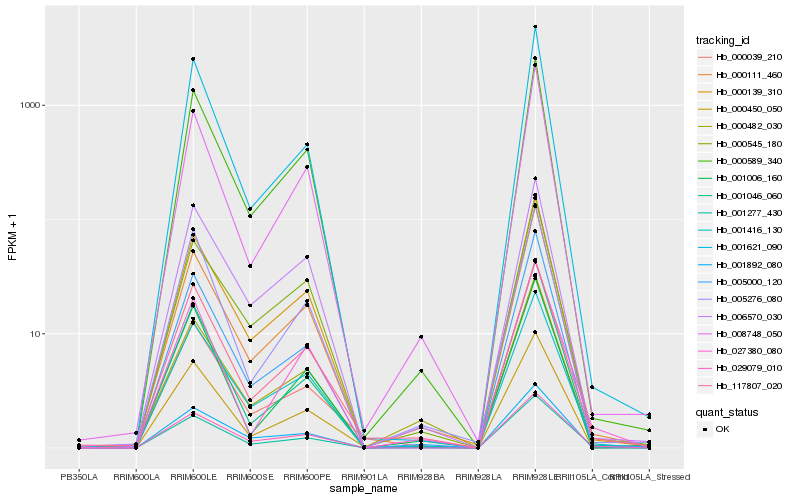
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001277_430 |
0.0 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 2 |
Hb_029079_010 |
0.0601650022 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 3 |
Hb_001416_130 |
0.062185713 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_117807_020 |
0.0793513859 |
- |
- |
methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_000589_340 |
0.0801198278 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 6 |
Hb_005000_120 |
0.0826537606 |
transcription factor |
TF Family: C2C2-GATA |
hypothetical protein RCOM_1046780 [Ricinus communis] |
| 7 |
Hb_000450_050 |
0.0840255009 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 8 |
Hb_000111_460 |
0.0878792021 |
- |
- |
PREDICTED: uncharacterized protein LOC105631266 [Jatropha curcas] |
| 9 |
Hb_000139_310 |
0.0884570129 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001621_090 |
0.0889854267 |
- |
- |
hypothetical protein POPTR_0010s20810g [Populus trichocarpa] |
| 11 |
Hb_027380_080 |
0.0916798547 |
- |
- |
hypothetical protein JCGZ_04649 [Jatropha curcas] |
| 12 |
Hb_008748_050 |
0.0926442071 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP24 10A, chloroplastic-like [Populus euphratica] |
| 13 |
Hb_005276_080 |
0.0932567445 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000039_210 |
0.0957550155 |
- |
- |
PREDICTED: uncharacterized protein LOC105649449 [Jatropha curcas] |
| 15 |
Hb_000545_180 |
0.0977354137 |
- |
- |
Photosystem II stability/assembly factor HCF136, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_001046_060 |
0.0988663815 |
- |
- |
PREDICTED: UDP-glucose flavonoid 3-O-glucosyltransferase 7-like [Fragaria vesca subsp. vesca] |
| 17 |
Hb_001892_080 |
0.1001723569 |
- |
- |
Clavata3/esr-related 16, putative [Theobroma cacao] |
| 18 |
Hb_006570_030 |
0.1006713257 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001006_160 |
0.1014090685 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 20 |
Hb_000482_030 |
0.1021537704 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like isoform X1 [Populus euphratica] |