| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
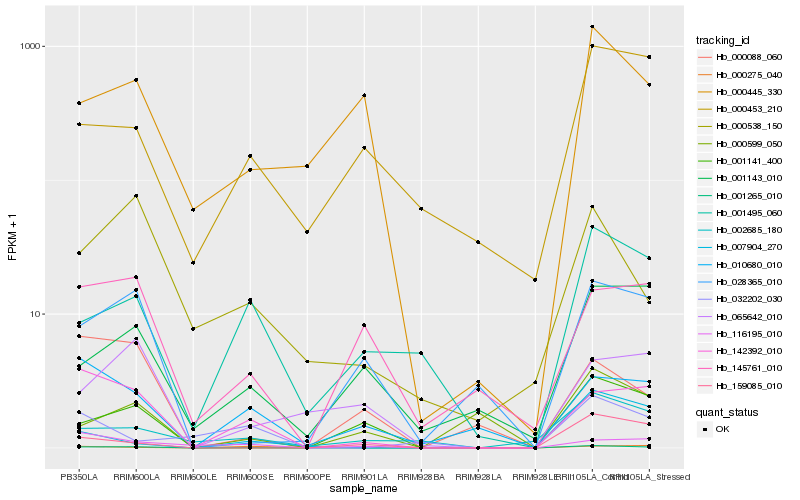
Hb_001265_010 |
0.0 |
transcription factor |
TF Family: NAC |
NAC transcription factor 057 [Jatropha curcas] |
| 2 |
Hb_142392_010 |
0.2389570499 |
- |
- |
PREDICTED: uncharacterized protein LOC105628741 [Jatropha curcas] |
| 3 |
Hb_001495_060 |
0.2743402025 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Jatropha curcas] |
| 4 |
Hb_010680_010 |
0.2936418431 |
- |
- |
- |
| 5 |
Hb_000538_150 |
0.2952167413 |
- |
- |
PREDICTED: 7-deoxyloganetin glucosyltransferase-like [Jatropha curcas] |
| 6 |
Hb_002685_180 |
0.2982007391 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_032202_030 |
0.3104865698 |
- |
- |
- |
| 8 |
Hb_007904_270 |
0.311144777 |
- |
- |
- |
| 9 |
Hb_028365_010 |
0.3122583163 |
- |
- |
PREDICTED: uncharacterized protein LOC105631661 [Jatropha curcas] |
| 10 |
Hb_001141_400 |
0.3141923008 |
- |
- |
hypothetical protein B456_007G2854002, partial [Gossypium raimondii] |
| 11 |
Hb_000445_330 |
0.3152381274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001143_010 |
0.3171320718 |
- |
- |
- |
| 13 |
Hb_000453_210 |
0.3210195142 |
- |
- |
hypothetical protein PHAVU_006G019800g [Phaseolus vulgaris] |
| 14 |
Hb_145761_010 |
0.3266235697 |
- |
- |
- |
| 15 |
Hb_116195_010 |
0.3310293142 |
- |
- |
- |
| 16 |
Hb_159085_010 |
0.3313804208 |
- |
- |
- |
| 17 |
Hb_000599_050 |
0.3331865265 |
- |
- |
vacuolar ATP synthase subunit G plant, putative [Ricinus communis] |
| 18 |
Hb_000275_040 |
0.3389488629 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 19 |
Hb_065642_010 |
0.3402858041 |
- |
- |
- |
| 20 |
Hb_000088_060 |
0.3409445578 |
- |
- |
- |