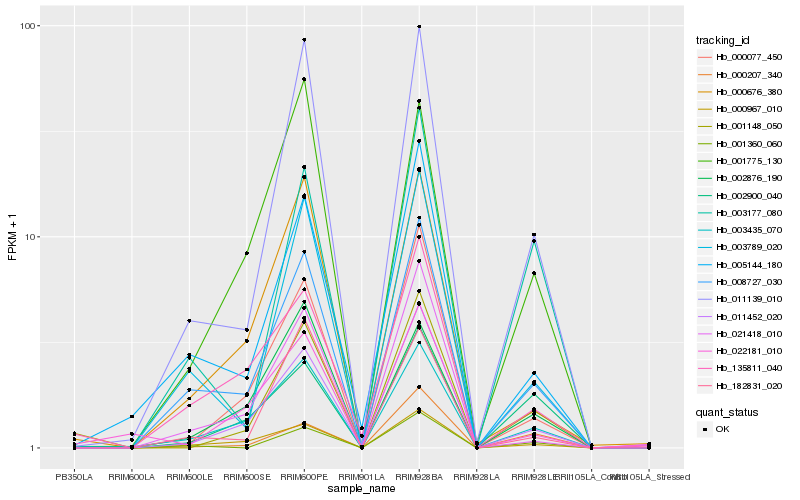
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001148_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_011452_020 |
0.1450688574 |
transcription factor |
TF Family: LOB |
PREDICTED: LOB domain-containing protein 4 [Jatropha curcas] |
| 3 |
Hb_000077_450 |
0.1503483703 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_011139_010 |
0.1533989443 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 5 |
Hb_003435_070 |
0.1719950194 |
- |
- |
PREDICTED: probable pectate lyase 12 isoform X3 [Jatropha curcas] |
| 6 |
Hb_008727_030 |
0.1797181637 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002900_040 |
0.1853520472 |
- |
- |
PREDICTED: 7-deoxyloganetic acid glucosyltransferase-like [Populus euphratica] |
| 8 |
Hb_000207_340 |
0.1902055286 |
transcription factor |
TF Family: GRAS |
hypothetical protein POPTR_0013s11930g [Populus trichocarpa] |
| 9 |
Hb_022181_010 |
0.1911641656 |
- |
- |
PREDICTED: peroxidase 19 [Jatropha curcas] |
| 10 |
Hb_000676_380 |
0.1924122707 |
- |
- |
cytochrome P450 family protein [Populus trichocarpa] |
| 11 |
Hb_003789_020 |
0.1936597424 |
- |
- |
hypothetical protein JCGZ_23602 [Jatropha curcas] |
| 12 |
Hb_003177_080 |
0.1968120392 |
- |
- |
hypothetical protein L484_016032 [Morus notabilis] |
| 13 |
Hb_000967_010 |
0.1971810312 |
- |
- |
hypothetical protein JCGZ_08336 [Jatropha curcas] |
| 14 |
Hb_001360_060 |
0.1987125725 |
- |
- |
PREDICTED: probable xyloglucan endotransglucosylase/hydrolase protein 32 [Jatropha curcas] |
| 15 |
Hb_005144_180 |
0.2006827595 |
- |
- |
PREDICTED: protein enabled homolog [Populus euphratica] |
| 16 |
Hb_001775_130 |
0.202641767 |
- |
- |
Prephenate dehydratase [Hevea brasiliensis] |
| 17 |
Hb_135811_040 |
0.2035436595 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_021418_010 |
0.2040812412 |
- |
- |
PREDICTED: probable pectate lyase 12 [Prunus mume] |
| 19 |
Hb_002876_190 |
0.2069789123 |
transcription factor |
TF Family: GRAS |
hypothetical protein PRUPE_ppa017519mg [Prunus persica] |
| 20 |
Hb_182831_020 |
0.2073108425 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |