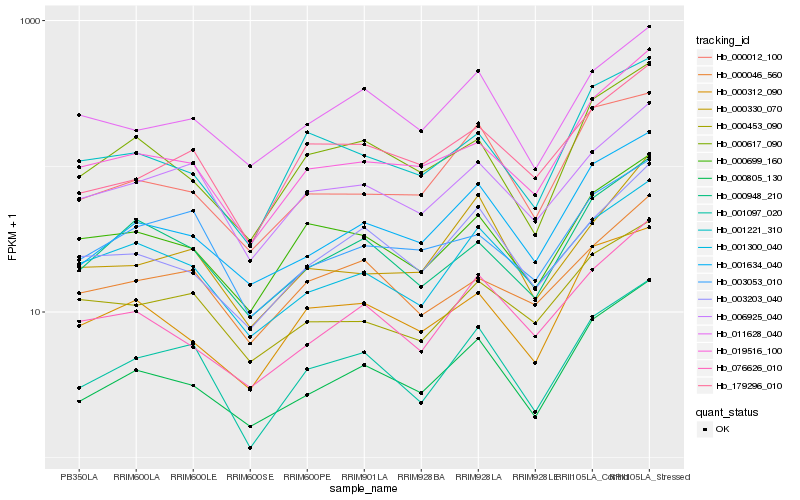
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001097_020 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s00490g [Populus trichocarpa] |
| 2 |
Hb_000805_130 |
0.1167742274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006925_040 |
0.1228240984 |
- |
- |
60S ribosomal protein L24-1 isoform 2 [Theobroma cacao] |
| 4 |
Hb_000948_210 |
0.1322905927 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA-like 2, mitochondrial [Jatropha curcas] |
| 5 |
Hb_001634_040 |
0.1323401343 |
transcription factor |
TF Family: NF-YC |
DNA polymerase epsilon subunit, putative [Ricinus communis] |
| 6 |
Hb_179296_010 |
0.1336562806 |
- |
- |
hypothetical protein B456_006G094200 [Gossypium raimondii] |
| 7 |
Hb_000699_160 |
0.1353212859 |
- |
- |
PREDICTED: C-Myc-binding protein homolog [Jatropha curcas] |
| 8 |
Hb_000617_090 |
0.1364126313 |
- |
- |
hypoia-responsive family protein 4 [Hevea brasiliensis] |
| 9 |
Hb_001300_040 |
0.1371810824 |
- |
- |
PREDICTED: uncharacterized protein LOC105642466 [Jatropha curcas] |
| 10 |
Hb_000330_070 |
0.1416317303 |
- |
- |
PREDICTED: proteasome subunit alpha type-1-B-like [Gossypium raimondii] |
| 11 |
Hb_001221_310 |
0.1428883611 |
- |
- |
sec61 gamma subunit, putative [Ricinus communis] |
| 12 |
Hb_003203_040 |
0.1429875998 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 7 [Vitis vinifera] |
| 13 |
Hb_019516_100 |
0.1430094534 |
- |
- |
40S ribosomal protein S14, putative [Ricinus communis] |
| 14 |
Hb_000312_090 |
0.1438178249 |
- |
- |
PREDICTED: uncharacterized protein LOC105645369 [Jatropha curcas] |
| 15 |
Hb_003053_010 |
0.1445636293 |
- |
- |
PREDICTED: uncharacterized protein LOC105641481 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000453_090 |
0.1445809525 |
- |
- |
mitochondrial import inner membrane translocase subunit Tim9 [Jatropha curcas] |
| 17 |
Hb_076626_010 |
0.1451414491 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_011628_040 |
0.1451940449 |
- |
- |
hypothetical protein CICLE_v10032980mg [Citrus clementina] |
| 19 |
Hb_000012_100 |
0.1460002928 |
- |
- |
UPF0587 protein C1orf123 homolog [Jatropha curcas] |
| 20 |
Hb_000046_560 |
0.1465812287 |
- |
- |
PREDICTED: uncharacterized protein LOC105118519 [Populus euphratica] |