| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
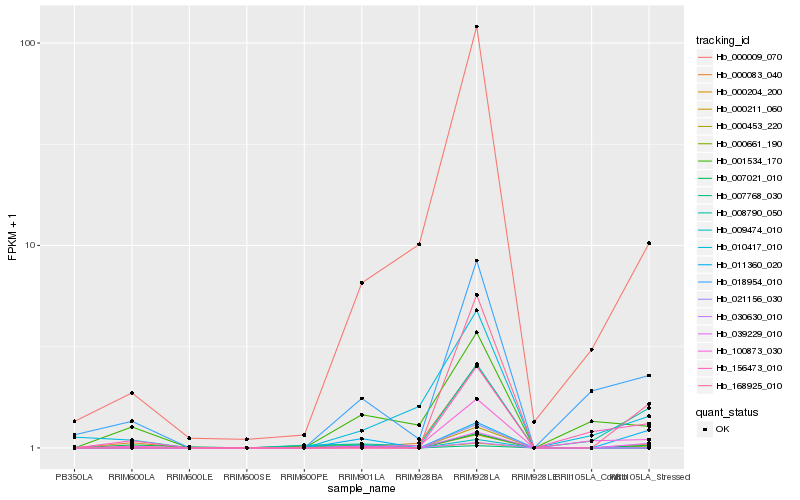
Hb_000453_220 |
0.0 |
- |
- |
hypothetical protein JCGZ_11079 [Jatropha curcas] |
| 2 |
Hb_018954_010 |
0.2445264506 |
- |
- |
- |
| 3 |
Hb_000009_070 |
0.2706312384 |
- |
- |
PREDICTED: globulin-1 S allele [Jatropha curcas] |
| 4 |
Hb_000083_040 |
0.2811791577 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007021_010 |
0.2820876333 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0939100 [Ricinus communis] |
| 6 |
Hb_001534_170 |
0.2845533504 |
- |
- |
Uncharacterized protein TCM_038456 [Theobroma cacao] |
| 7 |
Hb_000211_060 |
0.2859467883 |
- |
- |
glutathione peroxidase [Populus euphratica] |
| 8 |
Hb_000661_190 |
0.2876517001 |
- |
- |
PREDICTED: protein spt2-like [Jatropha curcas] |
| 9 |
Hb_168925_010 |
0.2891722422 |
- |
- |
- |
| 10 |
Hb_100873_030 |
0.2967139805 |
- |
- |
PREDICTED: uncharacterized protein LOC105795653 [Gossypium raimondii] |
| 11 |
Hb_007768_030 |
0.2998475403 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_009474_010 |
0.3027318648 |
- |
- |
- |
| 13 |
Hb_039229_010 |
0.3089908277 |
- |
- |
- |
| 14 |
Hb_000204_200 |
0.3113360155 |
- |
- |
hypothetical protein VITISV_019818 [Vitis vinifera] |
| 15 |
Hb_030630_010 |
0.3131264473 |
- |
- |
PREDICTED: uncharacterized protein LOC105786776 [Gossypium raimondii] |
| 16 |
Hb_011360_020 |
0.3144437417 |
- |
- |
- |
| 17 |
Hb_010417_010 |
0.317099476 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 18 |
Hb_021156_030 |
0.3192070099 |
- |
- |
hypothetical protein JCGZ_18422 [Jatropha curcas] |
| 19 |
Hb_008790_050 |
0.3214109737 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_156473_010 |
0.3218756485 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750 [Prunus mume] |