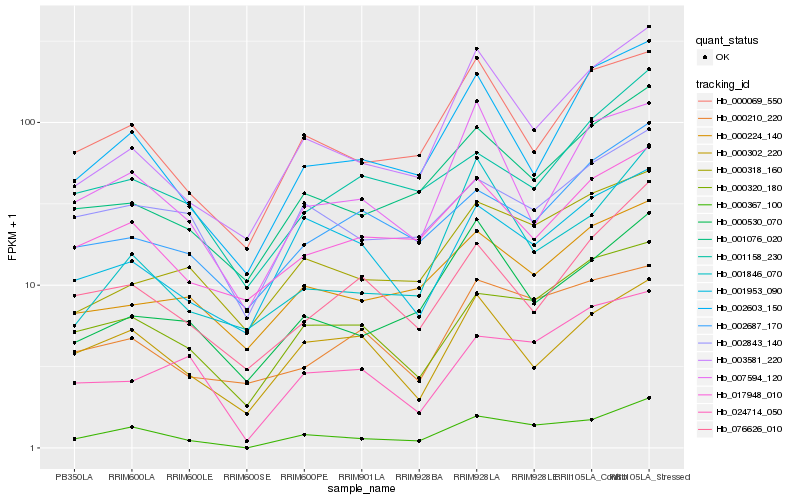
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000367_100 |
0.0 |
- |
- |
BnaA05g32070D [Brassica napus] |
| 2 |
Hb_000320_180 |
0.1404677493 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 3 |
Hb_003581_220 |
0.1407444756 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_001076_020 |
0.1452595052 |
- |
- |
PREDICTED: cytochrome b-c1 complex subunit 8 [Eucalyptus grandis] |
| 5 |
Hb_002603_150 |
0.1487312986 |
- |
- |
PREDICTED: protein yippee-like [Jatropha curcas] |
| 6 |
Hb_000069_550 |
0.1539383075 |
- |
- |
PREDICTED: uncharacterized protein LOC104596457 isoform X1 [Nelumbo nucifera] |
| 7 |
Hb_000318_160 |
0.1624462726 |
- |
- |
PREDICTED: uncharacterized protein LOC105634239 [Jatropha curcas] |
| 8 |
Hb_024714_050 |
0.1626058974 |
- |
- |
hypothetical protein CICLE_v100188992mg, partial [Citrus clementina] |
| 9 |
Hb_002843_140 |
0.1630938084 |
- |
- |
hypothetical protein JCGZ_20933 [Jatropha curcas] |
| 10 |
Hb_000530_070 |
0.1646870654 |
- |
- |
neutral/alkaline invertase 3 [Hevea brasiliensis] |
| 11 |
Hb_017948_010 |
0.1656601994 |
- |
- |
mitochondrial thioredoxin 2 [Hevea brasiliensis] |
| 12 |
Hb_000210_220 |
0.1682499789 |
- |
- |
hypothetical protein POPTR_0006s22010g [Populus trichocarpa] |
| 13 |
Hb_001953_090 |
0.1717518632 |
- |
- |
PREDICTED: membrane-anchored ubiquitin-fold protein 3 [Jatropha curcas] |
| 14 |
Hb_007594_120 |
0.1723327144 |
transcription factor |
TF Family: MIKC |
MADS-box transcription factor 2 [Hevea brasiliensis] |
| 15 |
Hb_076626_010 |
0.1724261282 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000224_140 |
0.1730071225 |
- |
- |
ribonuclease z, chloroplast, putative [Ricinus communis] |
| 17 |
Hb_001846_070 |
0.1731777652 |
- |
- |
PREDICTED: fructokinase-1 [Jatropha curcas] |
| 18 |
Hb_002687_170 |
0.1734057741 |
- |
- |
PREDICTED: G patch domain and ankyrin repeat-containing protein 1 homolog [Jatropha curcas] |
| 19 |
Hb_000302_220 |
0.174203407 |
- |
- |
PREDICTED: origin of replication complex subunit 5 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001158_230 |
0.1745793214 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM8 [Jatropha curcas] |