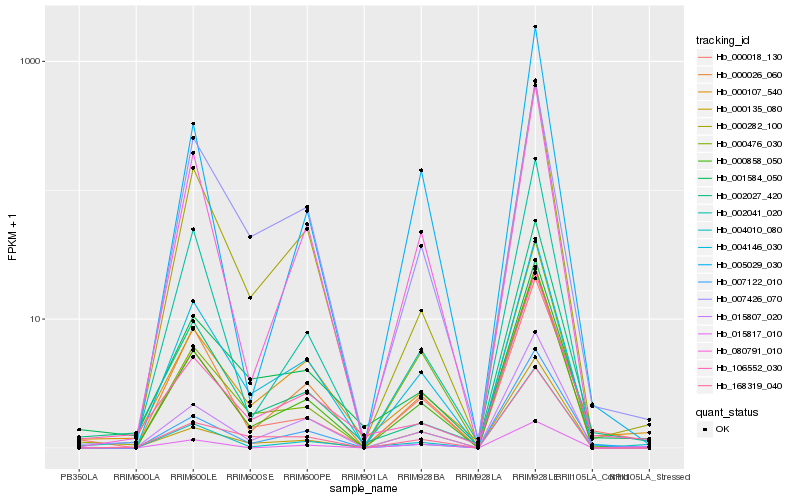
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000018_130 |
0.0 |
- |
- |
PREDICTED: probable lactoylglutathione lyase, chloroplast isoform X1 [Jatropha curcas] |
| 2 |
Hb_000858_050 |
0.1121723483 |
- |
- |
kinase, putative [Ricinus communis] |
| 3 |
Hb_004146_030 |
0.1333808321 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 4 |
Hb_005029_030 |
0.1411159763 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 36.4 kDa proline-rich protein-like [Sesamum indicum] |
| 5 |
Hb_000026_060 |
0.1417312379 |
- |
- |
beta-1,3-glucanase [Manihot esculenta] |
| 6 |
Hb_106552_030 |
0.1425215217 |
- |
- |
hypothetical protein POPTR_0006s23180g [Populus trichocarpa] |
| 7 |
Hb_004010_080 |
0.1469897429 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_080791_010 |
0.1471200723 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 9 |
Hb_002041_020 |
0.1490014743 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 10 |
Hb_007122_010 |
0.1519909071 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 11 |
Hb_015807_020 |
0.1530811614 |
- |
- |
gibberellin 20-oxidase, putative [Ricinus communis] |
| 12 |
Hb_000476_030 |
0.1534894496 |
- |
- |
PREDICTED: uncharacterized protein LOC105629366 [Jatropha curcas] |
| 13 |
Hb_002027_420 |
0.1541433054 |
- |
- |
hypothetical protein POPTR_0020s00240g [Populus trichocarpa] |
| 14 |
Hb_000282_100 |
0.15621486 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 15 |
Hb_007426_070 |
0.1585266927 |
- |
- |
PREDICTED: peroxisomal (S)-2-hydroxy-acid oxidase [Jatropha curcas] |
| 16 |
Hb_000135_080 |
0.1603090721 |
- |
- |
Flavonol synthase/flavanone 3-hydroxylase, putative [Ricinus communis] |
| 17 |
Hb_168319_040 |
0.1615037025 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Jatropha curcas] |
| 18 |
Hb_015817_010 |
0.1621260615 |
- |
- |
PREDICTED: putative receptor protein kinase ZmPK1 [Jatropha curcas] |
| 19 |
Hb_001584_050 |
0.1646084997 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000107_540 |
0.1646597059 |
- |
- |
PREDICTED: uncharacterized protein LOC105635335 [Jatropha curcas] |