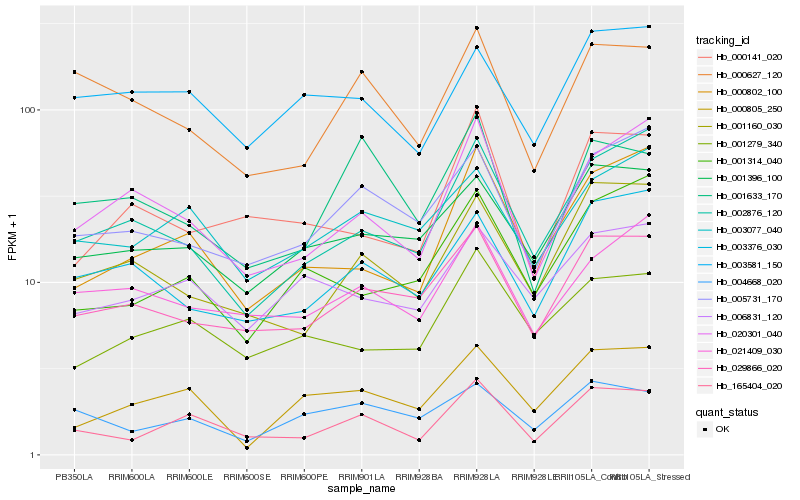
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_165404_020 |
0.0 |
- |
- |
hypothetical protein JCGZ_17789 [Jatropha curcas] |
| 2 |
Hb_000802_100 |
0.1377828219 |
- |
- |
PREDICTED: uncharacterized protein LOC105634009 [Jatropha curcas] |
| 3 |
Hb_029866_020 |
0.1431197726 |
- |
- |
PREDICTED: mitochondrial carrier protein MTM1 [Jatropha curcas] |
| 4 |
Hb_003077_040 |
0.1447207272 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase HIP1 [Jatropha curcas] |
| 5 |
Hb_005731_170 |
0.1466489961 |
- |
- |
hypothetical protein PRUPE_ppa011750mg [Prunus persica] |
| 6 |
Hb_001160_030 |
0.1482763857 |
transcription factor |
TF Family: Alfin-like |
hypothetical protein POPTR_0018s05470g [Populus trichocarpa] |
| 7 |
Hb_020301_040 |
0.1532920436 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein P [Jatropha curcas] |
| 8 |
Hb_001396_100 |
0.153948719 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_001279_340 |
0.1563772619 |
- |
- |
glycerol-3-phosphate dehydrogenase [Jatropha curcas] |
| 10 |
Hb_002876_120 |
0.1573230422 |
- |
- |
PREDICTED: protein-tyrosine-phosphatase PTP1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000627_120 |
0.1591404499 |
- |
- |
PREDICTED: probable adenylate kinase 7, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000141_020 |
0.1606460766 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 6 [Pyrus x bretschneideri] |
| 13 |
Hb_004668_020 |
0.1622494565 |
- |
- |
PREDICTED: uncharacterized protein LOC105630444 isoform X2 [Jatropha curcas] |
| 14 |
Hb_021409_030 |
0.1636442609 |
- |
- |
PREDICTED: serine/threonine-protein kinase WNK8 isoform X3 [Jatropha curcas] |
| 15 |
Hb_003581_150 |
0.1636969323 |
- |
- |
hypothetical protein CICLE_v10032366mg [Citrus clementina] |
| 16 |
Hb_000805_250 |
0.1637704377 |
- |
- |
PREDICTED: rhomboid-like protease 2 [Jatropha curcas] |
| 17 |
Hb_003376_030 |
0.1641325624 |
- |
- |
PREDICTED: ribonuclease P protein subunit p29 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001633_170 |
0.1646535448 |
- |
- |
PREDICTED: aspartokinase 2, chloroplastic-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_001314_040 |
0.1649504055 |
- |
- |
hypothetical protein POPTR_0011s10470g [Populus trichocarpa] |
| 20 |
Hb_006831_120 |
0.1650291775 |
- |
- |
PREDICTED: uncharacterized protein LOC105642121 isoform X1 [Jatropha curcas] |