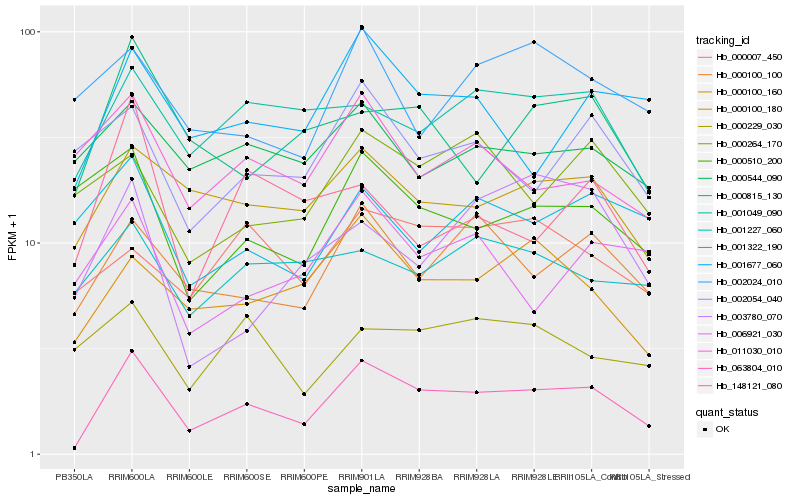
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_063804_010 |
0.0 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 2 |
Hb_000100_100 |
0.1776242694 |
- |
- |
hypothetical protein VITISV_027675 [Vitis vinifera] |
| 3 |
Hb_000100_160 |
0.1833298849 |
- |
- |
long-chain-fatty-acid CoA ligase, putative [Ricinus communis] |
| 4 |
Hb_000100_180 |
0.1849994762 |
- |
- |
PREDICTED: probable acyl-activating enzyme 16, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001677_060 |
0.1881933044 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit G-like [Jatropha curcas] |
| 6 |
Hb_001049_090 |
0.1956074983 |
- |
- |
6-phosphogluconate dehydrogenase, putative [Ricinus communis] |
| 7 |
Hb_000815_130 |
0.1987955771 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02-like [Jatropha curcas] |
| 8 |
Hb_006921_030 |
0.208076043 |
- |
- |
PREDICTED: uncharacterized protein LOC105638271 [Jatropha curcas] |
| 9 |
Hb_000007_450 |
0.2106991745 |
- |
- |
PREDICTED: tRNA(adenine(34)) deaminase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002054_040 |
0.2108757999 |
- |
- |
PREDICTED: putative clathrin assembly protein At5g35200 [Jatropha curcas] |
| 11 |
Hb_011030_010 |
0.2134948932 |
desease resistance |
Gene Name: RLI |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_000510_200 |
0.2135264862 |
- |
- |
nucleotide binding protein, putative [Ricinus communis] |
| 13 |
Hb_148121_080 |
0.214817025 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SHOOT GRAVITROPISM 5 [Populus euphratica] |
| 14 |
Hb_001322_190 |
0.2150172141 |
- |
- |
PREDICTED: uncharacterized protein LOC105632381 [Jatropha curcas] |
| 15 |
Hb_000264_170 |
0.2150303732 |
- |
- |
PREDICTED: uncharacterized protein LOC105649522 [Jatropha curcas] |
| 16 |
Hb_000544_090 |
0.2157291631 |
- |
- |
PREDICTED: golgin candidate 2 [Jatropha curcas] |
| 17 |
Hb_000229_030 |
0.2170486341 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g73710 [Jatropha curcas] |
| 18 |
Hb_001227_060 |
0.2174833572 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g20740 [Jatropha curcas] |
| 19 |
Hb_002024_010 |
0.2184843069 |
- |
- |
hypothetical protein B456_003G0564002, partial [Gossypium raimondii] |
| 20 |
Hb_003780_070 |
0.2203161233 |
- |
- |
PREDICTED: pirin-like protein [Jatropha curcas] |