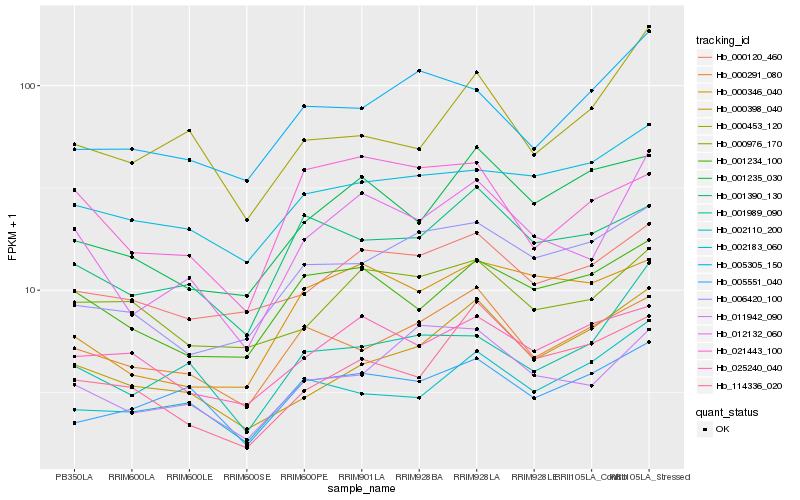
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012132_060 |
0.0 |
- |
- |
PREDICTED: centromere protein V [Jatropha curcas] |
| 2 |
Hb_001989_090 |
0.1235146103 |
- |
- |
PREDICTED: stomatin-like protein 2, mitochondrial [Jatropha curcas] |
| 3 |
Hb_011942_090 |
0.1288454164 |
- |
- |
PREDICTED: serine/threonine-protein kinase OSR1 isoform X1 [Jatropha curcas] |
| 4 |
Hb_001234_100 |
0.1319757405 |
- |
- |
PREDICTED: eyes absent homolog 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000120_460 |
0.1361375544 |
- |
- |
PREDICTED: thioredoxin reductase NTRB-like [Jatropha curcas] |
| 6 |
Hb_000976_170 |
0.1404341395 |
- |
- |
PREDICTED: ATPase family AAA domain-containing protein 3-like [Jatropha curcas] |
| 7 |
Hb_001390_130 |
0.1404503 |
- |
- |
hypothetical protein POPTR_0009s08590g [Populus trichocarpa] |
| 8 |
Hb_000346_040 |
0.1417127843 |
- |
- |
- |
| 9 |
Hb_002183_060 |
0.1432386759 |
transcription factor |
TF Family: C3H |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000398_040 |
0.1437739989 |
- |
- |
Auxin response factor, putative [Ricinus communis] |
| 11 |
Hb_001235_030 |
0.1456634442 |
- |
- |
PREDICTED: KH domain-containing protein At5g56140 [Vitis vinifera] |
| 12 |
Hb_000453_120 |
0.1457489376 |
- |
- |
PREDICTED: proteasome subunit alpha type-3 [Jatropha curcas] |
| 13 |
Hb_000291_080 |
0.1459424364 |
- |
- |
Vacuolar protein sorting-associated protein VPS9, putative [Ricinus communis] |
| 14 |
Hb_006420_100 |
0.1480476265 |
- |
- |
PREDICTED: protein bicaudal C homolog 1 [Jatropha curcas] |
| 15 |
Hb_005551_040 |
0.1491937676 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 16 |
Hb_002110_200 |
0.1494548255 |
- |
- |
PREDICTED: chaperone protein dnaJ 1, mitochondrial isoform X1 [Jatropha curcas] |
| 17 |
Hb_114336_020 |
0.1494870088 |
- |
- |
PREDICTED: cytosolic Fe-S cluster assembly factor narfl [Fragaria vesca subsp. vesca] |
| 18 |
Hb_025240_040 |
0.1511038079 |
- |
- |
hypothetical protein JCGZ_02438 [Jatropha curcas] |
| 19 |
Hb_021443_100 |
0.1511807196 |
- |
- |
- |
| 20 |
Hb_005305_150 |
0.1526205186 |
- |
- |
PREDICTED: 40S ribosomal protein S2-4-like [Jatropha curcas] |