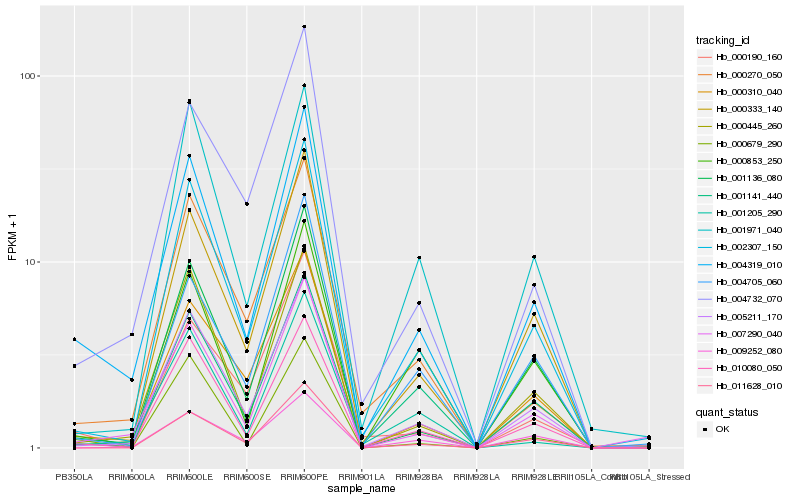
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004319_010 |
0.0 |
- |
- |
PREDICTED: peroxidase 63-like [Jatropha curcas] |
| 2 |
Hb_010080_050 |
0.1008155423 |
- |
- |
PREDICTED: uncharacterized protein LOC105635251 [Jatropha curcas] |
| 3 |
Hb_005211_170 |
0.1030834363 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 4 |
Hb_002307_150 |
0.1106755792 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: auxin-responsive protein IAA4-like [Jatropha curcas] |
| 5 |
Hb_007290_040 |
0.1107348824 |
- |
- |
PREDICTED: histidine-containing phosphotransfer protein 1-like [Jatropha curcas] |
| 6 |
Hb_000445_260 |
0.1178640163 |
- |
- |
PREDICTED: uncharacterized protein LOC105127729 [Populus euphratica] |
| 7 |
Hb_000333_140 |
0.1193175551 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 8 |
Hb_001136_080 |
0.1255015992 |
- |
- |
PREDICTED: protein ASPARTIC PROTEASE IN GUARD CELL 1 [Jatropha curcas] |
| 9 |
Hb_004732_070 |
0.1379329121 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_000679_290 |
0.1387951507 |
transcription factor |
TF Family: AUX/IAA |
ATP binding protein, putative [Ricinus communis] |
| 11 |
Hb_001141_440 |
0.1389447489 |
- |
- |
PREDICTED: uncharacterized protein LOC105631977 [Jatropha curcas] |
| 12 |
Hb_000310_040 |
0.1408115736 |
- |
- |
PREDICTED: adenine phosphoribosyltransferase 3 [Jatropha curcas] |
| 13 |
Hb_011628_010 |
0.1418560181 |
- |
- |
tonoplast intrinsic protein, putative [Ricinus communis] |
| 14 |
Hb_001205_290 |
0.1433838381 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP7-like [Jatropha curcas] |
| 15 |
Hb_000270_050 |
0.1437660227 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase FEI 2 [Jatropha curcas] |
| 16 |
Hb_004705_060 |
0.1462774025 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 17 |
Hb_000853_250 |
0.1520304683 |
- |
- |
Cellulose synthase A catalytic subunit 3 [UDP-forming], putative [Ricinus communis] |
| 18 |
Hb_001971_040 |
0.1522392173 |
- |
- |
PREDICTED: beta-glucosidase 44-like [Jatropha curcas] |
| 19 |
Hb_009252_080 |
0.1539319938 |
- |
- |
Polygalacturonase-1 non-catalytic subunit beta precursor, putative [Ricinus communis] |
| 20 |
Hb_000190_160 |
0.1539724455 |
- |
- |
PREDICTED: uncharacterized protein LOC105649940 isoform X2 [Jatropha curcas] |