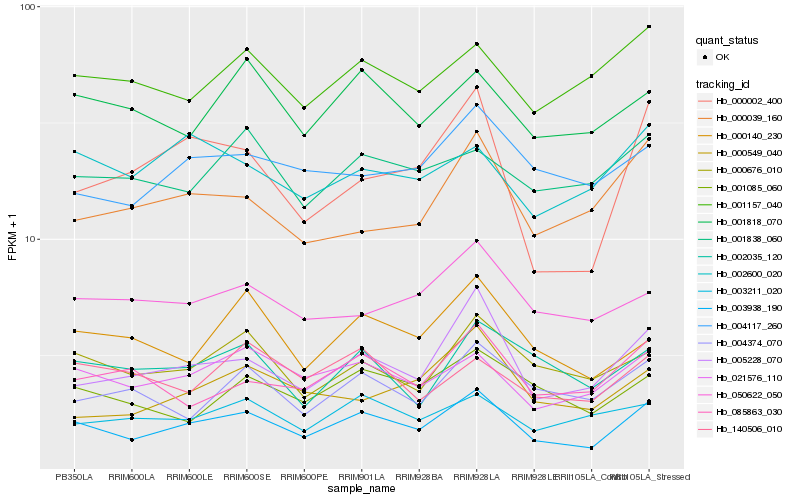
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003938_190 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g06145-like [Jatropha curcas] |
| 2 |
Hb_005228_070 |
0.0945640977 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g13230, mitochondrial [Jatropha curcas] |
| 3 |
Hb_021576_110 |
0.099509099 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 4 [Jatropha curcas] |
| 4 |
Hb_000676_010 |
0.1137157883 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g20230-like [Jatropha curcas] |
| 5 |
Hb_000002_400 |
0.1148784986 |
transcription factor |
TF Family: Orphans |
PREDICTED: ethylene receptor 2 [Jatropha curcas] |
| 6 |
Hb_001085_060 |
0.1198033402 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g04840 [Jatropha curcas] |
| 7 |
Hb_000140_230 |
0.1201419026 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61520, mitochondrial-like [Jatropha curcas] |
| 8 |
Hb_085863_030 |
0.120653225 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g31430 [Jatropha curcas] |
| 9 |
Hb_001818_070 |
0.1221250427 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 10 |
Hb_003211_020 |
0.1223889489 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g08310, mitochondrial [Jatropha curcas] |
| 11 |
Hb_001157_040 |
0.1233080203 |
- |
- |
casein kinase, putative [Ricinus communis] |
| 12 |
Hb_004117_260 |
0.1234760181 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002035_120 |
0.1254644977 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g14330 [Jatropha curcas] |
| 14 |
Hb_000039_160 |
0.1259798668 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase abkC [Jatropha curcas] |
| 15 |
Hb_004374_070 |
0.1263499539 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 16 |
Hb_050622_050 |
0.1270431566 |
- |
- |
PREDICTED: uncharacterized protein LOC105648353 [Jatropha curcas] |
| 17 |
Hb_140506_010 |
0.1281415274 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g33170-like [Jatropha curcas] |
| 18 |
Hb_000549_040 |
0.1287875236 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g12770-like isoform X1 [Jatropha curcas] |
| 19 |
Hb_002600_020 |
0.1294024835 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 20 |
Hb_001838_060 |
0.1295336814 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |