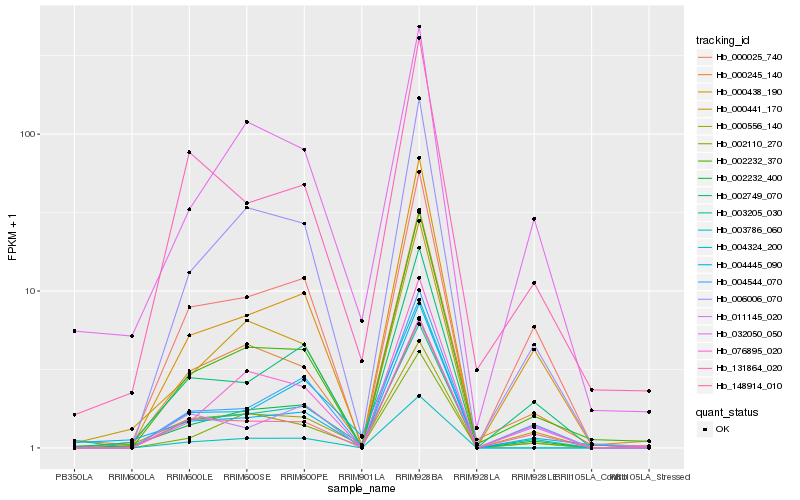
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003205_030 |
0.0 |
- |
- |
Pectinesterase inhibitor, putative [Ricinus communis] |
| 2 |
Hb_000556_140 |
0.1021791184 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 9 [Jatropha curcas] |
| 3 |
Hb_004544_070 |
0.1107029984 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 4 |
Hb_002232_400 |
0.1220909212 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH25-like [Jatropha curcas] |
| 5 |
Hb_002749_070 |
0.1229628362 |
- |
- |
PREDICTED: uncharacterized protein LOC105635941 [Jatropha curcas] |
| 6 |
Hb_006006_070 |
0.123995561 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_000438_190 |
0.1303470301 |
- |
- |
PREDICTED: peroxidase 4-like [Jatropha curcas] |
| 8 |
Hb_004445_090 |
0.1355291631 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_032050_050 |
0.136403575 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 10 |
Hb_002232_370 |
0.1377110392 |
- |
- |
PREDICTED: receptor-like protein kinase HAIKU2 [Jatropha curcas] |
| 11 |
Hb_000245_140 |
0.1394463517 |
- |
- |
PREDICTED: CASP-like protein 3A1 [Jatropha curcas] |
| 12 |
Hb_002110_270 |
0.1402616843 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH96 [Jatropha curcas] |
| 13 |
Hb_000025_740 |
0.1424134814 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000441_170 |
0.1439143638 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 15 |
Hb_131864_020 |
0.1458903875 |
- |
- |
PREDICTED: anthocyanidin reductase [Jatropha curcas] |
| 16 |
Hb_148914_010 |
0.1467158084 |
- |
- |
glutathione s-transferase, putative [Ricinus communis] |
| 17 |
Hb_003786_060 |
0.1467421222 |
- |
- |
PREDICTED: transmembrane protein 220 [Jatropha curcas] |
| 18 |
Hb_011145_020 |
0.1469783922 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Pyrus x bretschneideri] |
| 19 |
Hb_004324_200 |
0.1486565706 |
- |
- |
PREDICTED: transcription factor TRY-like [Populus euphratica] |
| 20 |
Hb_076895_020 |
0.1493016256 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X2 [Jatropha curcas] |