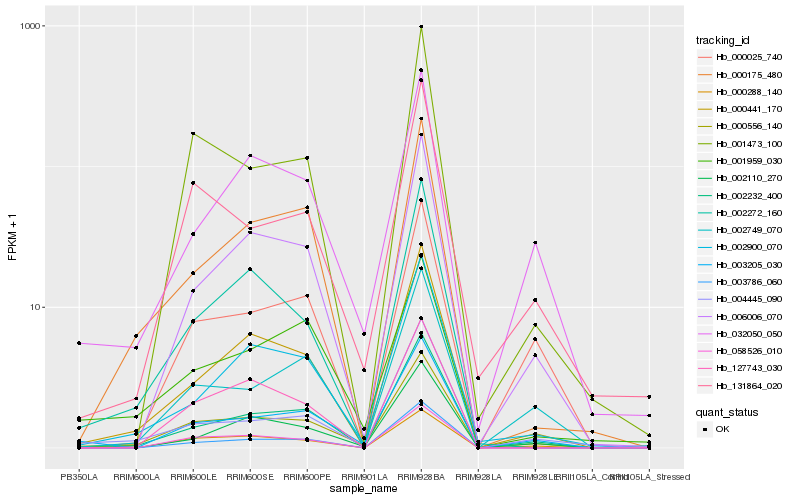
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000556_140 |
0.0 |
transcription factor |
TF Family: GRF |
PREDICTED: growth-regulating factor 9 [Jatropha curcas] |
| 2 |
Hb_003205_030 |
0.1021791184 |
- |
- |
Pectinesterase inhibitor, putative [Ricinus communis] |
| 3 |
Hb_000441_170 |
0.1077255159 |
- |
- |
PREDICTED: uncharacterized protein LOC105636851 [Jatropha curcas] |
| 4 |
Hb_002232_400 |
0.1131518529 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH25-like [Jatropha curcas] |
| 5 |
Hb_006006_070 |
0.1248748373 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000288_140 |
0.1249018056 |
- |
- |
PREDICTED: putative cysteine-rich receptor-like protein kinase 35 [Jatropha curcas] |
| 7 |
Hb_032050_050 |
0.1260656106 |
- |
- |
PREDICTED: peroxidase 12-like [Jatropha curcas] |
| 8 |
Hb_131864_020 |
0.1285630598 |
- |
- |
PREDICTED: anthocyanidin reductase [Jatropha curcas] |
| 9 |
Hb_002900_070 |
0.129225902 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002749_070 |
0.13781394 |
- |
- |
PREDICTED: uncharacterized protein LOC105635941 [Jatropha curcas] |
| 11 |
Hb_002272_160 |
0.1380659847 |
transcription factor |
TF Family: bHLH |
DNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_001473_100 |
0.138234057 |
- |
- |
chalcone synthase 1 [Jatropha curcas] |
| 13 |
Hb_000175_480 |
0.1385220154 |
- |
- |
hypothetical protein JCGZ_06165 [Jatropha curcas] |
| 14 |
Hb_058526_010 |
0.1401860886 |
- |
- |
- |
| 15 |
Hb_004445_090 |
0.1408769251 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_002110_270 |
0.1418887725 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH96 [Jatropha curcas] |
| 17 |
Hb_127743_030 |
0.1436778766 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 18 |
Hb_001959_030 |
0.1445249689 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_003786_060 |
0.1474260557 |
- |
- |
PREDICTED: transmembrane protein 220 [Jatropha curcas] |
| 20 |
Hb_000025_740 |
0.1486217988 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12 isoform X1 [Jatropha curcas] |