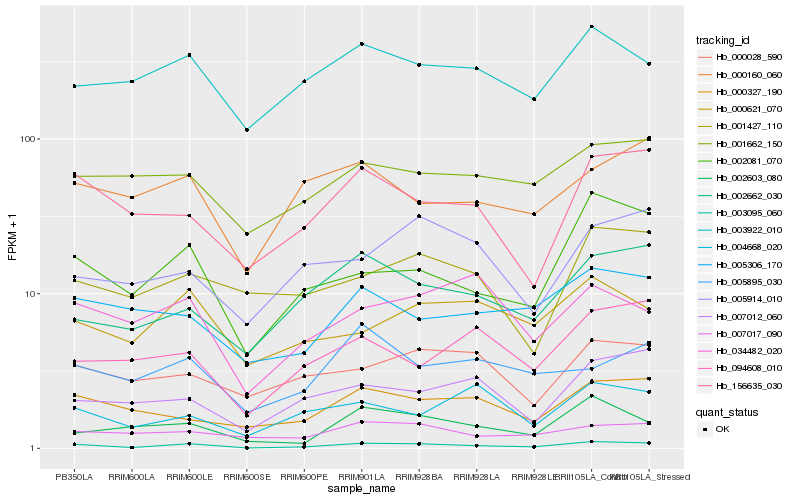
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003095_060 |
0.0 |
- |
- |
hypothetical protein VITISV_030841 [Vitis vinifera] |
| 2 |
Hb_002081_070 |
0.1571889583 |
- |
- |
PREDICTED: pyridoxal kinase [Jatropha curcas] |
| 3 |
Hb_156635_030 |
0.1668177836 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 8, mitochondrial isoform X1 [Nelumbo nucifera] |
| 4 |
Hb_002662_030 |
0.176863205 |
- |
- |
PREDICTED: mannose-P-dolichol utilization defect 1 protein homolog 2 [Jatropha curcas] |
| 5 |
Hb_003922_010 |
0.1804729206 |
- |
- |
PREDICTED: probable phospholipid hydroperoxide glutathione peroxidase [Jatropha curcas] |
| 6 |
Hb_000327_190 |
0.1808748345 |
- |
- |
PREDICTED: CDP-diacylglycerol--glycerol-3-phosphate 3-phosphatidyltransferase 2-like [Jatropha curcas] |
| 7 |
Hb_002603_080 |
0.1865844046 |
- |
- |
hypothetical protein MTR_7g114400 [Medicago truncatula] |
| 8 |
Hb_000621_070 |
0.1868957031 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 9 |
Hb_007017_090 |
0.1892231756 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 186 [Jatropha curcas] |
| 10 |
Hb_000028_590 |
0.1903069957 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Jatropha curcas] |
| 11 |
Hb_005306_170 |
0.1906658164 |
transcription factor |
TF Family: MYB-related |
PREDICTED: telomere repeat-binding factor 4-like [Jatropha curcas] |
| 12 |
Hb_001427_110 |
0.1912940532 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: yrdC domain-containing protein, mitochondrial [Jatropha curcas] |
| 13 |
Hb_007012_060 |
0.1913475818 |
- |
- |
PREDICTED: UPF0183 protein At3g51130 isoform X2 [Jatropha curcas] |
| 14 |
Hb_001662_150 |
0.1920088437 |
- |
- |
Scaffold attachment factor B1 [Medicago truncatula] |
| 15 |
Hb_005914_010 |
0.1956244933 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22 [Jatropha curcas] |
| 16 |
Hb_034482_020 |
0.1965421555 |
- |
- |
PREDICTED: bifunctional aspartokinase/homoserine dehydrogenase 1, chloroplastic-like [Jatropha curcas] |
| 17 |
Hb_000160_060 |
0.1967671073 |
- |
- |
unknown [Populus trichocarpa] |
| 18 |
Hb_004668_020 |
0.1970756754 |
- |
- |
PREDICTED: uncharacterized protein LOC105630444 isoform X2 [Jatropha curcas] |
| 19 |
Hb_005895_030 |
0.1976748033 |
- |
- |
PREDICTED: uncharacterized protein LOC105643388 [Jatropha curcas] |
| 20 |
Hb_094608_010 |
0.198294519 |
- |
- |
PREDICTED: uncharacterized protein LOC105630444 isoform X3 [Jatropha curcas] |