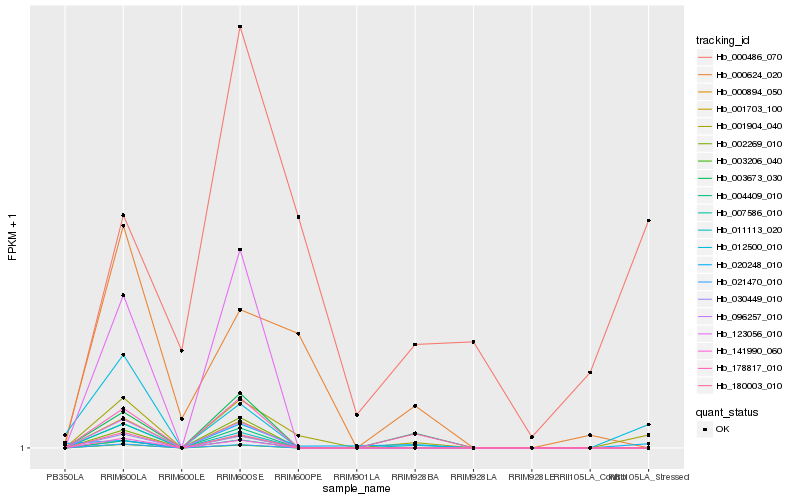
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001904_040 |
0.0 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 2 |
Hb_011113_020 |
0.326104161 |
- |
- |
hypothetical protein CICLE_v10018209mg [Citrus clementina] |
| 3 |
Hb_000624_020 |
0.3278684424 |
transcription factor |
TF Family: NAC |
NAC transcription factor 004 [Jatropha curcas] |
| 4 |
Hb_178817_010 |
0.341751284 |
- |
- |
protease [Gossypium hirsutum] |
| 5 |
Hb_012500_010 |
0.3486989837 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 6 |
Hb_000894_050 |
0.3501018439 |
- |
- |
- |
| 7 |
Hb_096257_010 |
0.3501282093 |
- |
- |
- |
| 8 |
Hb_180003_010 |
0.3505672748 |
- |
- |
PREDICTED: uncharacterized protein LOC105647154 [Jatropha curcas] |
| 9 |
Hb_030449_010 |
0.3506324259 |
- |
- |
- |
| 10 |
Hb_003673_030 |
0.3508798052 |
- |
- |
hypothetical protein POPTR_0011s11940g [Populus trichocarpa] |
| 11 |
Hb_021470_010 |
0.3510734479 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 12 |
Hb_004409_010 |
0.3514084553 |
- |
- |
- |
| 13 |
Hb_141990_060 |
0.3516221538 |
- |
- |
- |
| 14 |
Hb_003206_040 |
0.3517064543 |
- |
- |
Auxin efflux carrier component, putative [Ricinus communis] |
| 15 |
Hb_007586_010 |
0.351789223 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105126050 [Populus euphratica] |
| 16 |
Hb_020248_010 |
0.3523840769 |
- |
- |
PREDICTED: TMV resistance protein N-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_000486_070 |
0.3555734096 |
- |
- |
hypothetical protein JCGZ_25069 [Jatropha curcas] |
| 18 |
Hb_123056_010 |
0.3584039518 |
- |
- |
- |
| 19 |
Hb_001703_100 |
0.3665156763 |
- |
- |
PREDICTED: uncharacterized protein LOC104885230 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_002269_010 |
0.3684233642 |
- |
- |
PREDICTED: WAT1-related protein At4g15540-like [Jatropha curcas] |