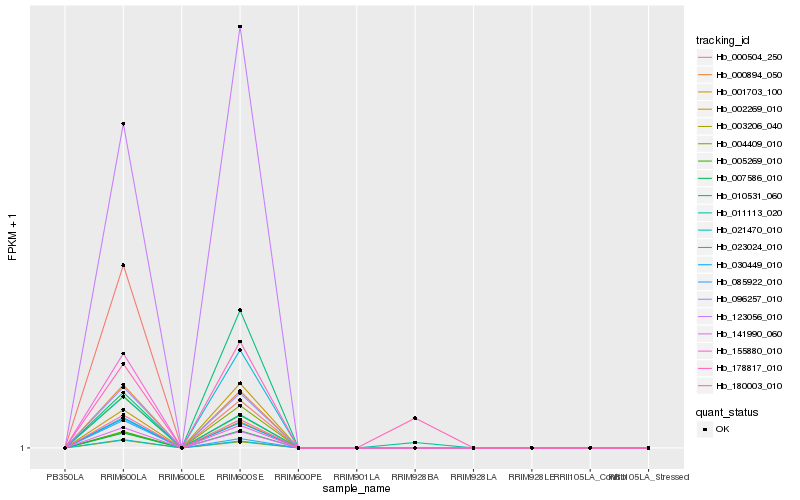
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_030449_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_180003_010 |
0.0010766053 |
- |
- |
PREDICTED: uncharacterized protein LOC105647154 [Jatropha curcas] |
| 3 |
Hb_021470_010 |
0.0063284333 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 4 |
Hb_096257_010 |
0.0102706378 |
- |
- |
- |
| 5 |
Hb_004409_010 |
0.01038778 |
- |
- |
- |
| 6 |
Hb_000894_050 |
0.0110156762 |
- |
- |
- |
| 7 |
Hb_141990_060 |
0.0127506123 |
- |
- |
- |
| 8 |
Hb_003206_040 |
0.0136431909 |
- |
- |
Auxin efflux carrier component, putative [Ricinus communis] |
| 9 |
Hb_007586_010 |
0.0641212426 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105126050 [Populus euphratica] |
| 10 |
Hb_123056_010 |
0.1079235078 |
- |
- |
- |
| 11 |
Hb_001703_100 |
0.1414468629 |
- |
- |
PREDICTED: uncharacterized protein LOC104885230 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_002269_010 |
0.1480916078 |
- |
- |
PREDICTED: WAT1-related protein At4g15540-like [Jatropha curcas] |
| 13 |
Hb_005269_010 |
0.178998886 |
- |
- |
- |
| 14 |
Hb_011113_020 |
0.1830452325 |
- |
- |
hypothetical protein CICLE_v10018209mg [Citrus clementina] |
| 15 |
Hb_085922_010 |
0.1878755081 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 16 |
Hb_010531_060 |
0.2322349315 |
- |
- |
hypothetical protein B456_010G209400, partial [Gossypium raimondii] |
| 17 |
Hb_000504_250 |
0.2498178484 |
- |
- |
- |
| 18 |
Hb_155880_010 |
0.2547348138 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 19 |
Hb_178817_010 |
0.2735016292 |
- |
- |
protease [Gossypium hirsutum] |
| 20 |
Hb_023024_010 |
0.276711194 |
- |
- |
PREDICTED: uncharacterized protein LOC105767094 [Gossypium raimondii] |