| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
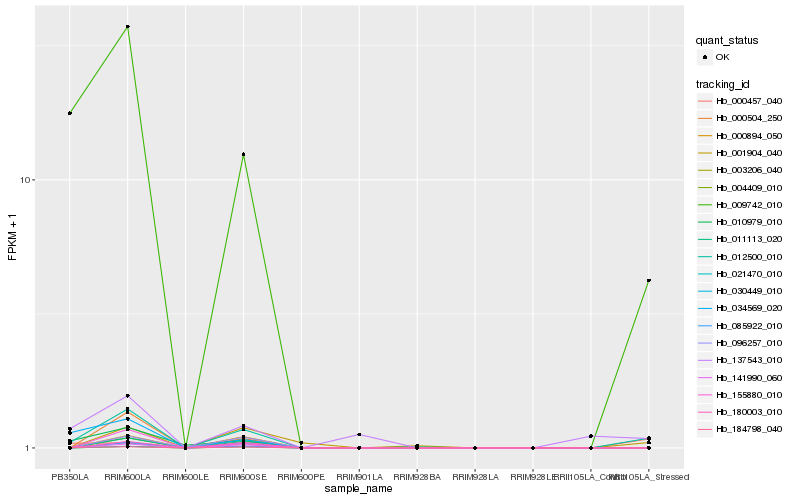
Hb_012500_010 |
0.0 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 2 |
Hb_009742_010 |
0.2379250357 |
- |
- |
- |
| 3 |
Hb_184798_040 |
0.3076478474 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 4 |
Hb_085922_010 |
0.3258941004 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 5 |
Hb_000504_250 |
0.3397581715 |
- |
- |
- |
| 6 |
Hb_137543_010 |
0.3410372415 |
- |
- |
- |
| 7 |
Hb_155880_010 |
0.341299462 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 8 |
Hb_003206_040 |
0.3434733753 |
- |
- |
Auxin efflux carrier component, putative [Ricinus communis] |
| 9 |
Hb_141990_060 |
0.3437623174 |
- |
- |
- |
| 10 |
Hb_004409_010 |
0.3445357352 |
- |
- |
- |
| 11 |
Hb_010979_010 |
0.3446401558 |
- |
- |
- |
| 12 |
Hb_021470_010 |
0.3458931615 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 13 |
Hb_034569_020 |
0.345893997 |
- |
- |
- |
| 14 |
Hb_030449_010 |
0.3480807502 |
- |
- |
- |
| 15 |
Hb_180003_010 |
0.348461459 |
- |
- |
PREDICTED: uncharacterized protein LOC105647154 [Jatropha curcas] |
| 16 |
Hb_001904_040 |
0.3486989837 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 17 |
Hb_096257_010 |
0.3518121207 |
- |
- |
- |
| 18 |
Hb_000894_050 |
0.3520913467 |
- |
- |
- |
| 19 |
Hb_011113_020 |
0.3675681878 |
- |
- |
hypothetical protein CICLE_v10018209mg [Citrus clementina] |
| 20 |
Hb_000457_040 |
0.3732994202 |
- |
- |
- |