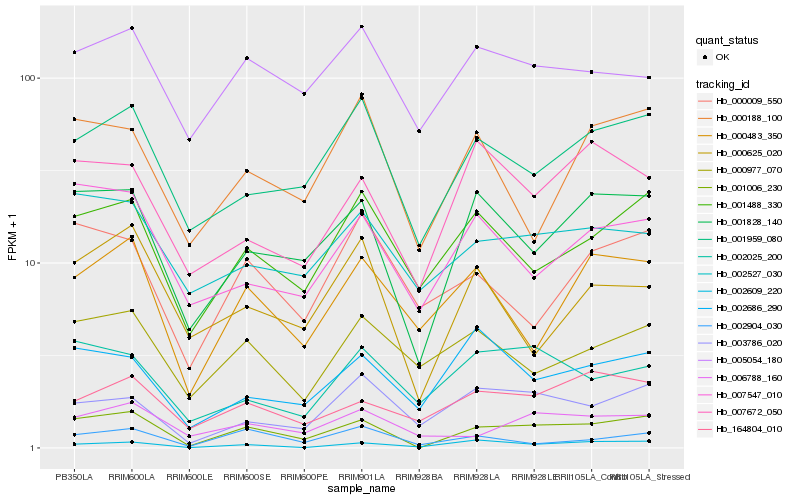
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001006_230 |
0.0 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 2 |
Hb_001828_140 |
0.1449028395 |
- |
- |
PREDICTED: uncharacterized protein LOC105633066 [Jatropha curcas] |
| 3 |
Hb_001959_080 |
0.1790443256 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001488_330 |
0.1855976794 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 8 [Jatropha curcas] |
| 5 |
Hb_002609_220 |
0.1967512299 |
- |
- |
Leucine-rich repeat protein kinase family protein, putative [Theobroma cacao] |
| 6 |
Hb_006788_160 |
0.1984261381 |
- |
- |
hypothetical protein POPTR_0002s04680g [Populus trichocarpa] |
| 7 |
Hb_007547_010 |
0.1987008965 |
- |
- |
PREDICTED: diphthamide biosynthesis protein 1 [Jatropha curcas] |
| 8 |
Hb_164804_010 |
0.1993072567 |
- |
- |
JHL23J11.8 [Jatropha curcas] |
| 9 |
Hb_002527_030 |
0.2013361415 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1-like [Jatropha curcas] |
| 10 |
Hb_003786_020 |
0.2040357945 |
- |
- |
PREDICTED: BTB/POZ domain-containing protein At3g50780 [Jatropha curcas] |
| 11 |
Hb_000483_350 |
0.204374893 |
- |
- |
PREDICTED: uncharacterized protein LOC105632381 [Jatropha curcas] |
| 12 |
Hb_002904_030 |
0.2046528129 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g71420 [Jatropha curcas] |
| 13 |
Hb_000625_020 |
0.2048311285 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000188_100 |
0.2055231257 |
- |
- |
PREDICTED: protein YIPF5 homolog [Jatropha curcas] |
| 15 |
Hb_007672_050 |
0.2062656586 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_005054_180 |
0.2072007733 |
- |
- |
coronatine-insensitive 1 [Hevea brasiliensis] |
| 17 |
Hb_002686_290 |
0.2074415557 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15010, mitochondrial [Populus euphratica] |
| 18 |
Hb_002025_200 |
0.2086072962 |
- |
- |
- |
| 19 |
Hb_000977_070 |
0.2088643613 |
- |
- |
PREDICTED: type II inositol 1,4,5-trisphosphate 5-phosphatase FRA3 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000009_550 |
0.2088645621 |
- |
- |
hypothetical protein RCOM_0603640 [Ricinus communis] |