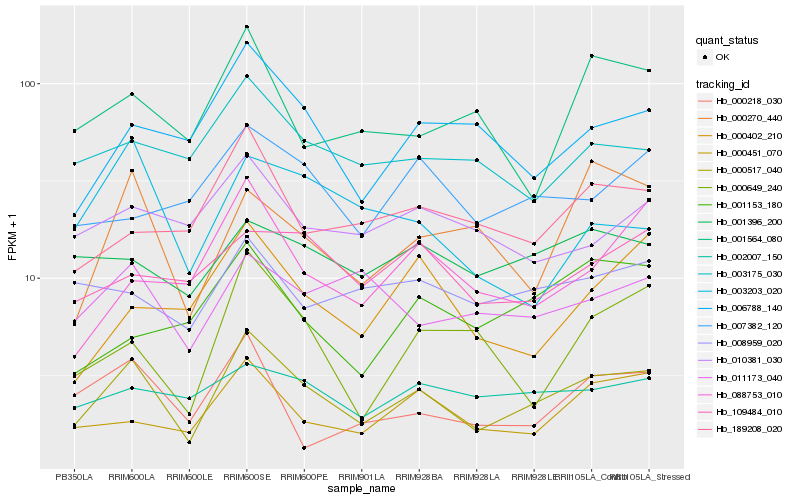
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000517_040 |
0.0 |
- |
- |
PREDICTED: protein argonaute 16 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000451_070 |
0.1690340915 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 3 |
Hb_006788_140 |
0.1716456806 |
- |
- |
PREDICTED: uncharacterized protein LOC105649915 [Jatropha curcas] |
| 4 |
Hb_189208_020 |
0.1784290466 |
- |
- |
LRX2, putative [Ricinus communis] |
| 5 |
Hb_000649_240 |
0.1805473381 |
- |
- |
PREDICTED: probable peroxygenase 4 [Jatropha curcas] |
| 6 |
Hb_000270_440 |
0.1811352987 |
transcription factor |
TF Family: Orphans |
hypothetical protein POPTR_0015s08130g [Populus trichocarpa] |
| 7 |
Hb_088753_010 |
0.1827276926 |
- |
- |
PREDICTED: copper amine oxidase 1-like [Vitis vinifera] |
| 8 |
Hb_003175_030 |
0.1829842787 |
- |
- |
PREDICTED: UBA and UBX domain-containing protein At4g15410-like [Populus euphratica] |
| 9 |
Hb_011173_040 |
0.1843466458 |
- |
- |
PREDICTED: protein ROOT PRIMORDIUM DEFECTIVE 1 [Populus euphratica] |
| 10 |
Hb_000218_030 |
0.1855770882 |
- |
- |
- |
| 11 |
Hb_002007_150 |
0.1875716888 |
- |
- |
- |
| 12 |
Hb_001396_200 |
0.1875886578 |
transcription factor |
TF Family: CAMTA |
PREDICTED: calmodulin-binding transcription activator 4 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001564_080 |
0.1880459855 |
transcription factor |
TF Family: NF-YB |
PREDICTED: protein Dr1 homolog isoform X2 [Jatropha curcas] |
| 14 |
Hb_000402_210 |
0.1882813511 |
- |
- |
hypothetical protein EUGRSUZ_B032971, partial [Eucalyptus grandis] |
| 15 |
Hb_007382_120 |
0.1896278626 |
- |
- |
PREDICTED: delta(3,5)-Delta(2,4)-dienoyl-CoA isomerase, mitochondrial [Jatropha curcas] |
| 16 |
Hb_008959_020 |
0.1896511747 |
- |
- |
PREDICTED: nitric oxide synthase-interacting protein [Pyrus x bretschneideri] |
| 17 |
Hb_010381_030 |
0.1905142017 |
- |
- |
PREDICTED: uncharacterized protein LOC105631461 [Jatropha curcas] |
| 18 |
Hb_001153_180 |
0.1923820085 |
- |
- |
PREDICTED: SPX domain-containing membrane protein At4g22990-like [Jatropha curcas] |
| 19 |
Hb_109484_010 |
0.1928055869 |
- |
- |
PREDICTED: INO80 complex subunit D-like [Jatropha curcas] |
| 20 |
Hb_003203_020 |
0.1930635737 |
- |
- |
unknown [Lotus japonicus] |