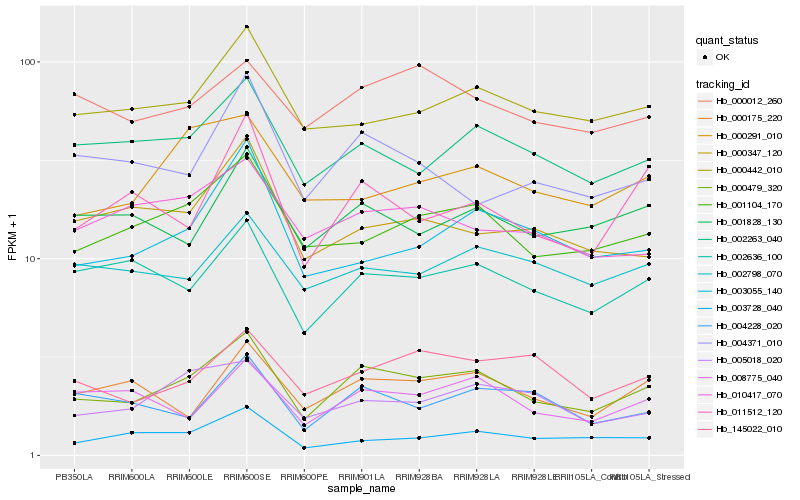
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000479_320 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g06140, mitochondrial-like [Jatropha curcas] |
| 2 |
Hb_001104_170 |
0.1112091311 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor A-2c [Jatropha curcas] |
| 3 |
Hb_002263_040 |
0.1153487302 |
- |
- |
carbon catabolite repressor protein, putative [Ricinus communis] |
| 4 |
Hb_002636_100 |
0.1196717678 |
- |
- |
PREDICTED: DNA-directed RNA polymerase 1B, mitochondrial [Jatropha curcas] |
| 5 |
Hb_005018_020 |
0.1240680714 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g07740, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000442_010 |
0.1245988848 |
- |
- |
hypothetical protein POPTR_0010s19060g [Populus trichocarpa] |
| 7 |
Hb_011512_120 |
0.1270990376 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 1 [Jatropha curcas] |
| 8 |
Hb_145022_010 |
0.1289873972 |
- |
- |
hypothetical protein JCGZ_22699 [Jatropha curcas] |
| 9 |
Hb_008775_040 |
0.1290862686 |
- |
- |
Pentatricopeptide repeat (PPR) superfamily protein, putative [Theobroma cacao] |
| 10 |
Hb_001828_130 |
0.1295632398 |
- |
- |
PREDICTED: transcription factor GTE8-like [Jatropha curcas] |
| 11 |
Hb_003055_140 |
0.1314814995 |
transcription factor |
TF Family: SNF2 |
PREDICTED: uncharacterized protein LOC105649495 [Jatropha curcas] |
| 12 |
Hb_010417_070 |
0.1316448302 |
- |
- |
PREDICTED: uncharacterized protein LOC105645744 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000012_260 |
0.1319738141 |
- |
- |
Far upstream element-binding 2 [Gossypium arboreum] |
| 14 |
Hb_000175_220 |
0.1329690155 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g21065-like [Jatropha curcas] |
| 15 |
Hb_002798_070 |
0.1331649236 |
transcription factor |
TF Family: Jumonji |
PREDICTED: putative lysine-specific demethylase JMJ16 [Jatropha curcas] |
| 16 |
Hb_000291_010 |
0.1343537327 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase 1 [Jatropha curcas] |
| 17 |
Hb_003728_040 |
0.1355089542 |
- |
- |
PREDICTED: uncharacterized protein LOC101492305 [Cicer arietinum] |
| 18 |
Hb_004228_020 |
0.1358429545 |
- |
- |
hypothetical protein JCGZ_21866 [Jatropha curcas] |
| 19 |
Hb_004371_010 |
0.1361944778 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1b(c38)-like isoform X3 [Jatropha curcas] |
| 20 |
Hb_000347_120 |
0.1368450712 |
- |
- |
PREDICTED: uncharacterized protein LOC105631363 [Jatropha curcas] |