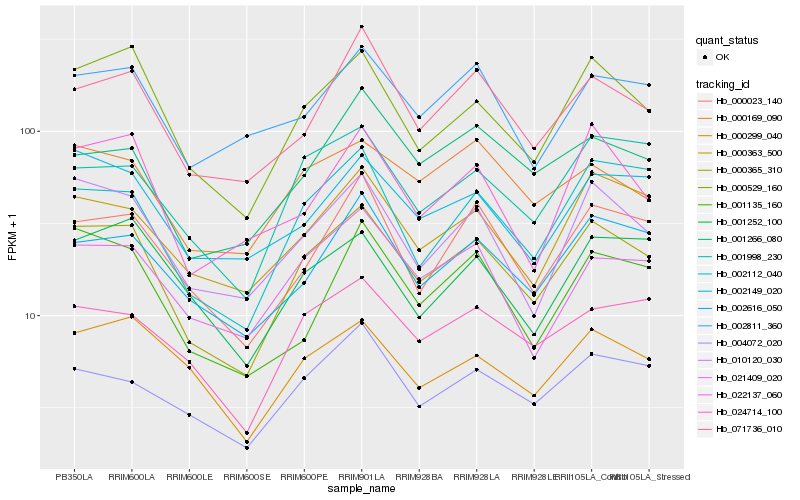
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000365_310 |
0.0 |
- |
- |
auxin:hydrogen symporter, putative [Ricinus communis] |
| 2 |
Hb_000529_160 |
0.0724166837 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Populus euphratica] |
| 3 |
Hb_010120_030 |
0.0846052231 |
- |
- |
PREDICTED: heterogeneous nuclear ribonucleoprotein 1 [Jatropha curcas] |
| 4 |
Hb_002112_040 |
0.0919406723 |
- |
- |
PREDICTED: uncharacterized protein LOC105641009 [Jatropha curcas] |
| 5 |
Hb_000363_500 |
0.0931538025 |
- |
- |
PREDICTED: EH domain-containing protein 1 isoform X2 [Jatropha curcas] |
| 6 |
Hb_002616_050 |
0.0941454237 |
- |
- |
PREDICTED: traB domain-containing protein [Jatropha curcas] |
| 7 |
Hb_001998_230 |
0.0946283983 |
- |
- |
PREDICTED: coatomer subunit epsilon-1 [Jatropha curcas] |
| 8 |
Hb_000023_140 |
0.0962958029 |
- |
- |
PREDICTED: cadmium-induced protein AS8 isoform X1 [Jatropha curcas] |
| 9 |
Hb_071736_010 |
0.0971597928 |
- |
- |
Metallopeptidase M24 family protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_024714_100 |
0.0976154929 |
- |
- |
PREDICTED: alkylated DNA repair protein alkB homolog 8 isoform X1 [Jatropha curcas] |
| 11 |
Hb_004072_020 |
0.0987582483 |
- |
- |
PREDICTED: uncharacterized protein LOC105635274 [Jatropha curcas] |
| 12 |
Hb_021409_020 |
0.1016032992 |
- |
- |
PREDICTED: protein yippee-like At4g27745 [Malus domestica] |
| 13 |
Hb_000169_090 |
0.1028226853 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like [Jatropha curcas] |
| 14 |
Hb_000299_040 |
0.1030458515 |
- |
- |
PREDICTED: protein S-acyltransferase 10 isoform X1 [Jatropha curcas] |
| 15 |
Hb_002149_020 |
0.1036171775 |
- |
- |
PREDICTED: uncharacterized protein LOC105647982 isoform X2 [Jatropha curcas] |
| 16 |
Hb_022137_060 |
0.1082816438 |
- |
- |
1-acylglycerophosphocholine O-acyltransferase [Vernicia fordii] |
| 17 |
Hb_002811_360 |
0.1086891749 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 18 |
Hb_001135_160 |
0.1087363937 |
- |
- |
PREDICTED: elongator complex protein 6 [Jatropha curcas] |
| 19 |
Hb_001266_080 |
0.1094522286 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 2 subunit gamma-like [Jatropha curcas] |
| 20 |
Hb_001252_100 |
0.1097761181 |
- |
- |
PREDICTED: CASP-like protein 5B2 [Jatropha curcas] |