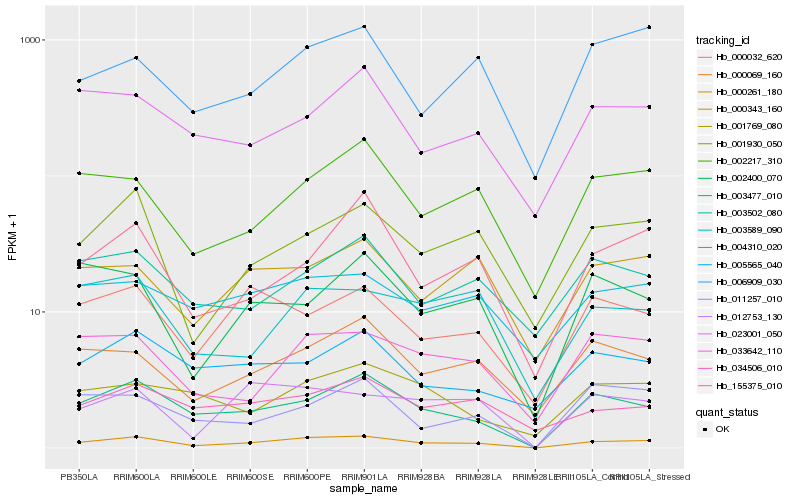
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_180 |
0.0 |
- |
- |
Tetratricopeptide repeat-like superfamily protein, putative isoform 1 [Theobroma cacao] |
| 2 |
Hb_003477_010 |
0.1142791686 |
- |
- |
hypothetical protein JCGZ_21107 [Jatropha curcas] |
| 3 |
Hb_001930_050 |
0.149567569 |
- |
- |
Serine acetyltransferase 3, mitochondrial precursor, putative [Ricinus communis] |
| 4 |
Hb_000069_160 |
0.1562305247 |
- |
- |
- |
| 5 |
Hb_002217_310 |
0.1564006057 |
- |
- |
PREDICTED: ER membrane protein complex subunit 6 [Jatropha curcas] |
| 6 |
Hb_033642_110 |
0.1590196212 |
- |
- |
PREDICTED: MLO-like protein 4 [Jatropha curcas] |
| 7 |
Hb_012753_130 |
0.1593283565 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_155375_010 |
0.1634073951 |
- |
- |
hypothetical protein JCGZ_23388 [Jatropha curcas] |
| 9 |
Hb_000343_160 |
0.1658694477 |
- |
- |
PREDICTED: protein SYS1 homolog [Jatropha curcas] |
| 10 |
Hb_000032_620 |
0.1658899039 |
- |
- |
hypothetical protein RCOM_1311830 [Ricinus communis] |
| 11 |
Hb_034506_010 |
0.1660997312 |
- |
- |
PREDICTED: protein OS-9 homolog [Jatropha curcas] |
| 12 |
Hb_004310_020 |
0.1696147538 |
- |
- |
PREDICTED: COP9 signalosome complex subunit 7 isoform X1 [Jatropha curcas] |
| 13 |
Hb_003589_090 |
0.1696559345 |
- |
- |
PREDICTED: potassium transporter 2 [Jatropha curcas] |
| 14 |
Hb_006909_030 |
0.1710585746 |
- |
- |
40S ribosomal protein S17B [Hevea brasiliensis] |
| 15 |
Hb_023001_050 |
0.1736376444 |
- |
- |
hypothetical protein POPTR_0006s10330g [Populus trichocarpa] |
| 16 |
Hb_003502_080 |
0.1742076612 |
- |
- |
vacuolar protein sorting, putative [Ricinus communis] |
| 17 |
Hb_002400_070 |
0.1742862776 |
transcription factor |
TF Family: AUX/IAA |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g56140 [Jatropha curcas] |
| 18 |
Hb_005565_040 |
0.1761283617 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_011257_010 |
0.1765394683 |
- |
- |
- |
| 20 |
Hb_001769_080 |
0.1770123626 |
- |
- |
hypothetical protein SORBIDRAFT_10g023200 [Sorghum bicolor] |