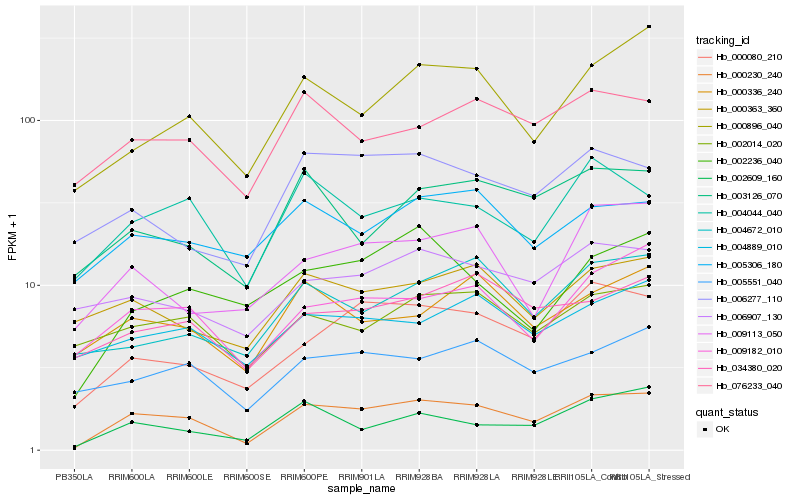
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000230_240 |
0.0 |
- |
- |
tRNA/rRNA methyltransferase family protein [Populus trichocarpa] |
| 2 |
Hb_000080_210 |
0.1384085499 |
- |
- |
PREDICTED: uncharacterized protein LOC105628422 [Jatropha curcas] |
| 3 |
Hb_002609_160 |
0.144884859 |
- |
- |
PREDICTED: GDP-mannose transporter GONST2 [Jatropha curcas] |
| 4 |
Hb_009182_010 |
0.1455079851 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 20 homolog 2-like [Jatropha curcas] |
| 5 |
Hb_004044_040 |
0.1486075283 |
- |
- |
PREDICTED: NADH-cytochrome b5 reductase-like protein [Jatropha curcas] |
| 6 |
Hb_000896_040 |
0.1507025716 |
- |
- |
BnaC02g13240D [Brassica napus] |
| 7 |
Hb_076233_040 |
0.1566489044 |
- |
- |
PREDICTED: ATP synthase subunit gamma, mitochondrial [Jatropha curcas] |
| 8 |
Hb_002236_040 |
0.158317691 |
- |
- |
PREDICTED: cytochrome c [Jatropha curcas] |
| 9 |
Hb_003126_070 |
0.1585227477 |
- |
- |
PREDICTED: 3-dehydroquinate synthase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_005551_040 |
0.1592518921 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 11 |
Hb_009113_050 |
0.1605569509 |
- |
- |
PREDICTED: uncharacterized protein LOC105644587 [Jatropha curcas] |
| 12 |
Hb_006277_110 |
0.1609612997 |
- |
- |
protein with unknown function [Ricinus communis] |
| 13 |
Hb_000336_240 |
0.1619828845 |
transcription factor |
TF Family: mTERF |
LOC100285792 [Zea mays] |
| 14 |
Hb_034380_020 |
0.1623786611 |
- |
- |
PREDICTED: phosphoglycerate mutase-like [Jatropha curcas] |
| 15 |
Hb_002014_020 |
0.1643727005 |
- |
- |
PREDICTED: protein Mpv17 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000363_360 |
0.1661787699 |
- |
- |
hypothetical protein PRUPE_ppa009146mg [Prunus persica] |
| 17 |
Hb_005306_180 |
0.1663695927 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 2 of pyruvate dehydrogenase complex, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_004889_010 |
0.1667011761 |
- |
- |
Vacuolar protein sorting-associated protein-like protein [Medicago truncatula] |
| 19 |
Hb_004672_010 |
0.1667207965 |
- |
- |
ecotropic viral integration site, putative [Ricinus communis] |
| 20 |
Hb_006907_130 |
0.1670324225 |
- |
- |
unnamed protein product [Coffea canephora] |