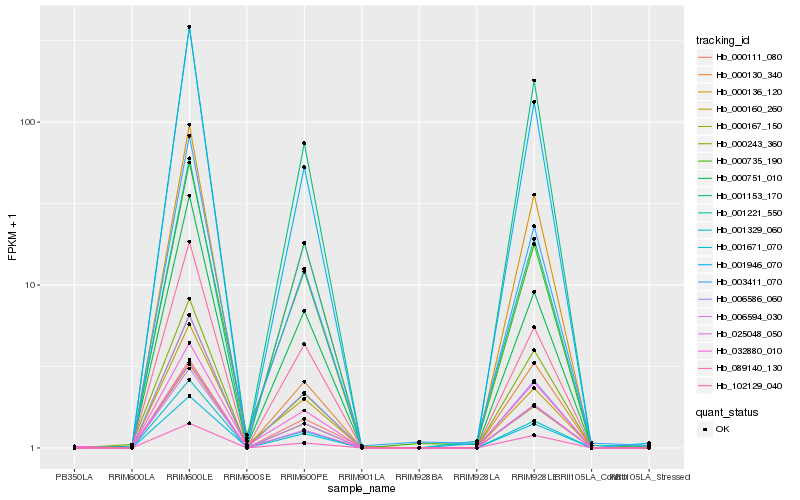
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000111_080 |
0.0 |
- |
- |
hypothetical protein JCGZ_24534 [Jatropha curcas] |
| 2 |
Hb_000136_120 |
0.0855513708 |
- |
- |
PREDICTED: probable strigolactone esterase D14 homolog [Jatropha curcas] |
| 3 |
Hb_000167_150 |
0.0857275319 |
- |
- |
hypothetical protein POPTR_0008s03590g [Populus trichocarpa] |
| 4 |
Hb_006586_060 |
0.0874728359 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |
| 5 |
Hb_006594_030 |
0.0902271294 |
- |
- |
- |
| 6 |
Hb_102129_040 |
0.0931972819 |
- |
- |
PREDICTED: PTI1-like tyrosine-protein kinase 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000735_190 |
0.0950578025 |
- |
- |
hypothetical protein POPTR_0006s19340g [Populus trichocarpa] |
| 8 |
Hb_001329_060 |
0.0987603743 |
- |
- |
Polygalacturonase precursor, putative [Ricinus communis] |
| 9 |
Hb_001946_070 |
0.0989552633 |
transcription factor |
TF Family: ERF |
DNA binding protein, putative [Ricinus communis] |
| 10 |
Hb_000130_340 |
0.0998930148 |
- |
- |
- |
| 11 |
Hb_000243_360 |
0.1001371691 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 7 isoform X1 [Vitis vinifera] |
| 12 |
Hb_032880_010 |
0.1001467521 |
- |
- |
PREDICTED: beta-glucosidase 24-like [Jatropha curcas] |
| 13 |
Hb_001221_550 |
0.1013656581 |
- |
- |
Corticosteroid 11-beta-dehydrogenase, putative [Ricinus communis] |
| 14 |
Hb_003411_070 |
0.102182665 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 3.1 [Populus euphratica] |
| 15 |
Hb_000751_010 |
0.1028902801 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_001153_170 |
0.1041517959 |
- |
- |
PREDICTED: protein HOTHEAD [Populus euphratica] |
| 17 |
Hb_001671_070 |
0.1054908857 |
- |
- |
kinase, putative [Ricinus communis] |
| 18 |
Hb_000160_260 |
0.1059887066 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 19 |
Hb_025048_050 |
0.1080725847 |
- |
- |
PREDICTED: protein PHYTOCHROME KINASE SUBSTRATE 3 [Jatropha curcas] |
| 20 |
Hb_089140_130 |
0.1091731457 |
- |
- |
hypothetical protein POPTR_0006s14510g [Populus trichocarpa] |