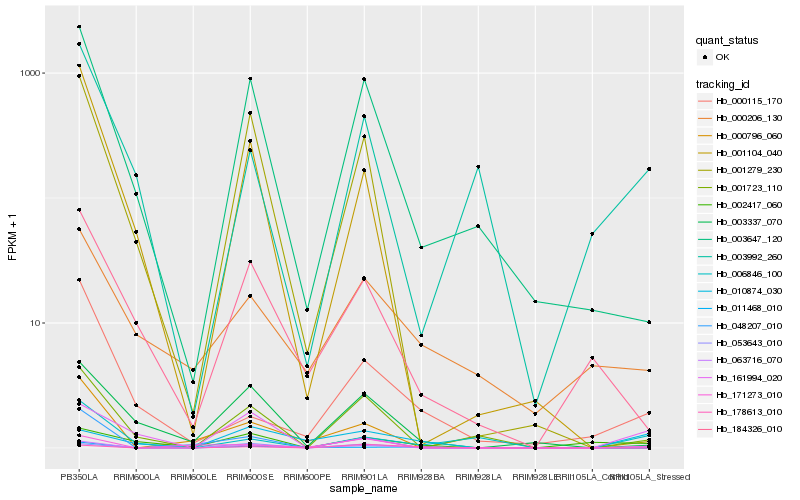
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_063716_070 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa020565mg [Prunus persica] |
| 2 |
Hb_001723_110 |
0.2940758693 |
- |
- |
PREDICTED: solute carrier family 25 member 44-like isoform X2 [Malus domestica] |
| 3 |
Hb_003647_120 |
0.2941971319 |
- |
- |
18.2 kDa class I heat shock family protein [Populus trichocarpa] |
| 4 |
Hb_006846_100 |
0.3104424321 |
- |
- |
hypothetical protein POPTR_0012s11270g [Populus trichocarpa] |
| 5 |
Hb_161994_020 |
0.3156428694 |
- |
- |
- |
| 6 |
Hb_001279_230 |
0.3161733683 |
- |
- |
PREDICTED: class I heat shock protein-like [Jatropha curcas] |
| 7 |
Hb_003337_070 |
0.3189370133 |
- |
- |
PREDICTED: probable esterase KAI2 [Jatropha curcas] |
| 8 |
Hb_010874_030 |
0.3256487995 |
transcription factor |
TF Family: M-type |
hypothetical protein JCGZ_10946 [Jatropha curcas] |
| 9 |
Hb_003992_260 |
0.3310013529 |
- |
- |
PREDICTED: 26.5 kDa heat shock protein, mitochondrial [Jatropha curcas] |
| 10 |
Hb_002417_060 |
0.3326944121 |
- |
- |
Cell division cycle protein, putative [Ricinus communis] |
| 11 |
Hb_000206_130 |
0.3332596824 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 12 |
Hb_011468_010 |
0.3363546973 |
- |
- |
hypothetical protein B456_008G198600 [Gossypium raimondii] |
| 13 |
Hb_048207_010 |
0.3410644948 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 14 |
Hb_184326_010 |
0.3414947749 |
- |
- |
RecName: Full=Anthocyanidin 3-O-glucosyltransferase 1; AltName: Full=Flavonol 3-O-glucosyltransferase 1; AltName: Full=UDP-glucose flavonoid 3-O-glucosyltransferase 1 [Manihot esculenta] |
| 15 |
Hb_001104_040 |
0.3431527636 |
- |
- |
PREDICTED: uncharacterized protein LOC105629509 [Jatropha curcas] |
| 16 |
Hb_171273_010 |
0.3434266559 |
- |
- |
E3 ubiquitin-protein ligase UPL5 [Theobroma cacao] |
| 17 |
Hb_000115_170 |
0.3478465106 |
- |
- |
hypothetical protein JCGZ_18339 [Jatropha curcas] |
| 18 |
Hb_053643_010 |
0.3486113813 |
- |
- |
Zinc finger protein GIS2 [Medicago truncatula] |
| 19 |
Hb_178613_010 |
0.3505174532 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 20 |
Hb_000796_060 |
0.352962867 |
- |
- |
Glycogenin-2 [Aegilops tauschii] |