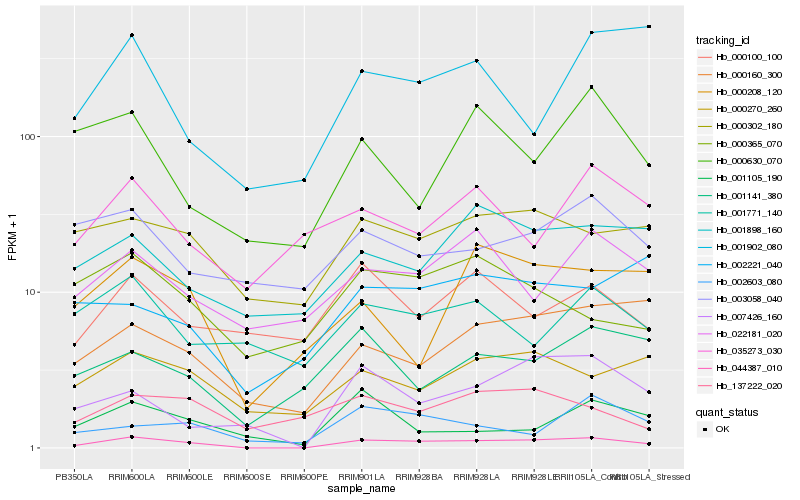
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_044387_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 2 |
Hb_007426_160 |
0.2254995595 |
- |
- |
- |
| 3 |
Hb_000160_300 |
0.2335665777 |
- |
- |
hypothetical protein POPTR_0004s08720g [Populus trichocarpa] |
| 4 |
Hb_000302_180 |
0.2394717396 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 5 |
Hb_000270_260 |
0.2435390259 |
- |
- |
PREDICTED: uncharacterized protein LOC105640466 [Jatropha curcas] |
| 6 |
Hb_022181_020 |
0.2510514817 |
- |
- |
PREDICTED: D-tyrosyl-tRNA(Tyr) deacylase isoform X1 [Jatropha curcas] |
| 7 |
Hb_001898_160 |
0.2513112397 |
- |
- |
hypothetical protein POPTR_0004s23390g [Populus trichocarpa] |
| 8 |
Hb_001141_380 |
0.252083867 |
- |
- |
hypothetical protein PRUPE_ppa012823mg [Prunus persica] |
| 9 |
Hb_002603_080 |
0.2520900648 |
- |
- |
hypothetical protein MTR_7g114400 [Medicago truncatula] |
| 10 |
Hb_000208_120 |
0.2523546594 |
- |
- |
PREDICTED: CAP-Gly domain-containing linker protein 1-like [Jatropha curcas] |
| 11 |
Hb_001105_190 |
0.2624610755 |
- |
- |
peptidyl-prolyl cis-trans isomerase, putative [Ricinus communis] |
| 12 |
Hb_000100_100 |
0.2627568745 |
- |
- |
hypothetical protein VITISV_027675 [Vitis vinifera] |
| 13 |
Hb_000630_070 |
0.2629548962 |
- |
- |
PREDICTED: cell division protein FtsZ homolog 2-1, chloroplastic [Jatropha curcas] |
| 14 |
Hb_137222_020 |
0.2636012792 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 15 |
Hb_001771_140 |
0.2641240242 |
- |
- |
PREDICTED: protein HIRA isoform X1 [Jatropha curcas] |
| 16 |
Hb_002221_040 |
0.2645066052 |
- |
- |
PREDICTED: uncharacterized protein LOC105639851 [Jatropha curcas] |
| 17 |
Hb_000365_070 |
0.2648688995 |
- |
- |
PREDICTED: SWR1 complex subunit 2 [Jatropha curcas] |
| 18 |
Hb_003058_040 |
0.2650524869 |
- |
- |
rotamase, putative [Ricinus communis] |
| 19 |
Hb_001902_080 |
0.2655387534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_035273_030 |
0.2655433699 |
rubber biosynthesis |
Gene Name: Mevalonate kinase |
mevalonate kinase [Hevea brasiliensis] |