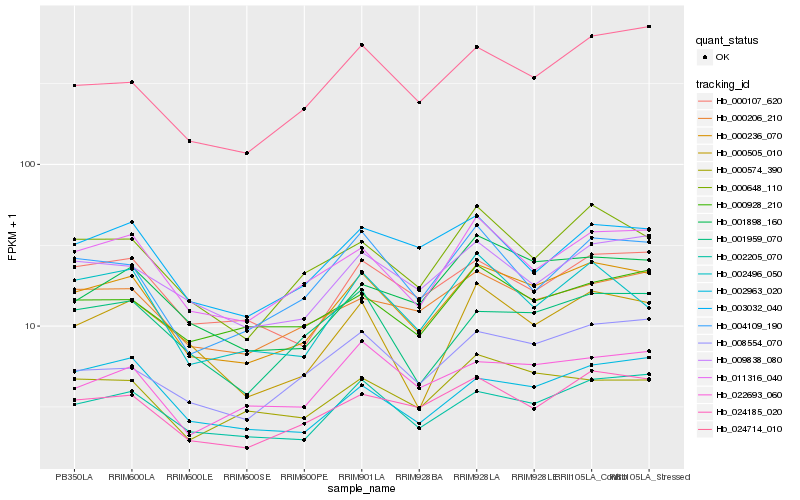
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000236_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_002205_070 |
0.0685845297 |
- |
- |
PREDICTED: tRNA (guanine(26)-N(2))-dimethyltransferase isoform X1 [Jatropha curcas] |
| 3 |
Hb_011316_040 |
0.0721327472 |
- |
- |
peptidase, putative [Ricinus communis] |
| 4 |
Hb_024185_020 |
0.0776902846 |
- |
- |
- |
| 5 |
Hb_000107_620 |
0.0825891272 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15 [Jatropha curcas] |
| 6 |
Hb_000648_110 |
0.0830393757 |
- |
- |
PREDICTED: D-xylose-proton symporter-like 3, chloroplastic [Jatropha curcas] |
| 7 |
Hb_009838_080 |
0.0868765363 |
- |
- |
Ras-GTPase-activating protein-binding protein, putative [Ricinus communis] |
| 8 |
Hb_000206_210 |
0.0900319597 |
- |
- |
PREDICTED: G patch domain-containing protein 11 [Jatropha curcas] |
| 9 |
Hb_004109_190 |
0.0908405804 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 25-like isoform X2 [Populus euphratica] |
| 10 |
Hb_024714_010 |
0.0923627923 |
- |
- |
HSP20-like chaperones superfamily protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_001898_160 |
0.0935992269 |
- |
- |
hypothetical protein POPTR_0004s23390g [Populus trichocarpa] |
| 12 |
Hb_002496_050 |
0.0937452811 |
- |
- |
PREDICTED: uncharacterized protein LOC105630509 [Jatropha curcas] |
| 13 |
Hb_001959_070 |
0.0940000534 |
- |
- |
PREDICTED: uncharacterized protein LOC105638451 isoform X1 [Jatropha curcas] |
| 14 |
Hb_002963_020 |
0.0942442976 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_022693_060 |
0.0947337109 |
- |
- |
- |
| 16 |
Hb_000574_390 |
0.0950931697 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 6-like [Jatropha curcas] |
| 17 |
Hb_008554_070 |
0.0961534468 |
- |
- |
PREDICTED: peroxisome biogenesis protein 3-2 isoform X2 [Jatropha curcas] |
| 18 |
Hb_003032_040 |
0.09675268 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: zinc finger CCCH domain-containing protein 25 [Jatropha curcas] |
| 19 |
Hb_000505_010 |
0.0993082282 |
- |
- |
PREDICTED: uncharacterized protein LOC105641242 [Jatropha curcas] |
| 20 |
Hb_000928_210 |
0.0997439502 |
- |
- |
PREDICTED: uncharacterized protein LOC105632512 isoform X1 [Jatropha curcas] |