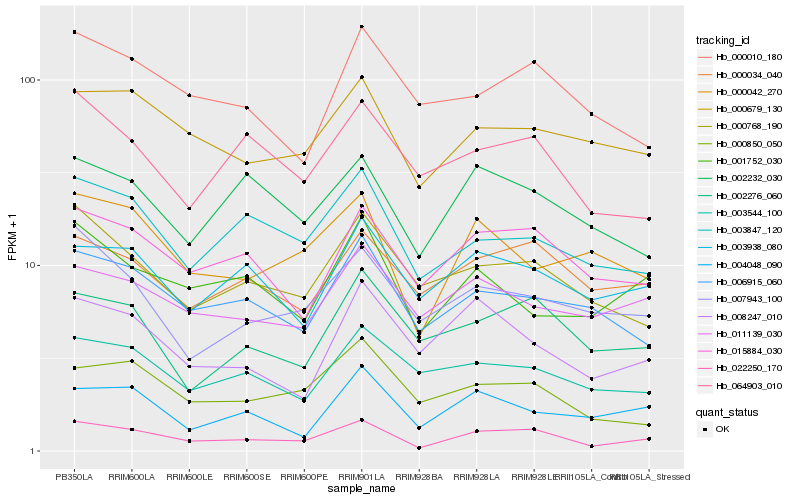
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_022250_170 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g26782, mitochondrial-like [Populus euphratica] |
| 2 |
Hb_015884_030 |
0.1363792648 |
- |
- |
PREDICTED: uncharacterized protein LOC105647778 isoform X1 [Jatropha curcas] |
| 3 |
Hb_003847_120 |
0.1480832702 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 4 |
Hb_000042_270 |
0.1492008957 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 5 |
Hb_000679_130 |
0.1492248472 |
- |
- |
dead box ATP-dependent RNA helicase, putative [Ricinus communis] |
| 6 |
Hb_000034_040 |
0.1515040214 |
- |
- |
PREDICTED: histidine--tRNA ligase [Jatropha curcas] |
| 7 |
Hb_000768_190 |
0.1523773844 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 8 |
Hb_006915_060 |
0.1536096068 |
- |
- |
PREDICTED: zinc finger BED domain-containing protein DAYSLEEPER-like [Jatropha curcas] |
| 9 |
Hb_002276_060 |
0.1546601795 |
- |
- |
hypothetical protein CISIN_1g002019mg [Citrus sinensis] |
| 10 |
Hb_064903_010 |
0.155132428 |
- |
- |
PREDICTED: T-complex protein 1 subunit delta [Jatropha curcas] |
| 11 |
Hb_001752_030 |
0.1554068302 |
- |
- |
PREDICTED: RNA polymerase II C-terminal domain phosphatase-like 4 [Jatropha curcas] |
| 12 |
Hb_002232_030 |
0.155498947 |
- |
- |
Pinin, putative [Ricinus communis] |
| 13 |
Hb_003938_080 |
0.1562299793 |
- |
- |
PREDICTED: uncharacterized protein LOC105644554 [Jatropha curcas] |
| 14 |
Hb_007943_100 |
0.1569266742 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_008247_010 |
0.1576336035 |
- |
- |
PREDICTED: leucine--tRNA ligase, cytoplasmic [Jatropha curcas] |
| 16 |
Hb_000850_050 |
0.1579294336 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like [Jatropha curcas] |
| 17 |
Hb_004048_090 |
0.1604401471 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g53330 [Jatropha curcas] |
| 18 |
Hb_011139_030 |
0.1622532362 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase MBR2-like [Jatropha curcas] |
| 19 |
Hb_000010_180 |
0.1642346544 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic-like [Jatropha curcas] |
| 20 |
Hb_003544_100 |
0.1647821999 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g59900 [Jatropha curcas] |