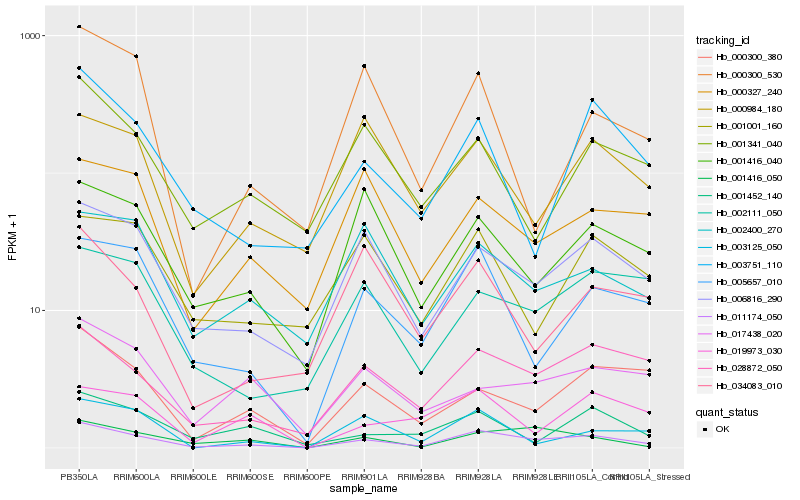
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011174_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_028872_050 |
0.1736586321 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] flavoprotein 1, mitochondrial [Jatropha curcas] |
| 3 |
Hb_006816_290 |
0.175389581 |
- |
- |
PREDICTED: uncharacterized protein LOC105647449 [Jatropha curcas] |
| 4 |
Hb_005657_010 |
0.1847339235 |
- |
- |
- |
| 5 |
Hb_001416_050 |
0.2028766516 |
- |
- |
PREDICTED: uncharacterized protein LOC104107076 [Nicotiana tomentosiformis] |
| 6 |
Hb_000300_380 |
0.2048249066 |
- |
- |
- |
| 7 |
Hb_001416_040 |
0.2099955894 |
- |
- |
acid phosphatase, putative [Ricinus communis] |
| 8 |
Hb_003751_110 |
0.2100258694 |
- |
- |
unknown [Lotus japonicus] |
| 9 |
Hb_003125_050 |
0.2135972381 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g50390, chloroplastic [Jatropha curcas] |
| 10 |
Hb_002400_270 |
0.2180367887 |
- |
- |
PREDICTED: probable sucrose-phosphate synthase 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_002111_050 |
0.218876834 |
- |
- |
PREDICTED: uncharacterized protein LOC105641242 [Jatropha curcas] |
| 12 |
Hb_000327_240 |
0.2195393909 |
- |
- |
PREDICTED: interaptin-like [Jatropha curcas] |
| 13 |
Hb_001341_040 |
0.2232597242 |
- |
- |
hypothetical protein JCGZ_05578 [Jatropha curcas] |
| 14 |
Hb_017438_020 |
0.2241083933 |
- |
- |
- |
| 15 |
Hb_000984_180 |
0.2245591671 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO3 isoform X1 [Jatropha curcas] |
| 16 |
Hb_001452_140 |
0.2258126579 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Elaeis guineensis] |
| 17 |
Hb_034083_010 |
0.2261663441 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_019973_030 |
0.2279063022 |
- |
- |
- |
| 19 |
Hb_001001_160 |
0.2283167961 |
- |
- |
Phosphatidic acid phosphohydrolase 2 isoform 1 [Theobroma cacao] |
| 20 |
Hb_000300_530 |
0.2296517691 |
- |
- |
PREDICTED: uncharacterized protein DDB_G0286299-like [Gossypium raimondii] |