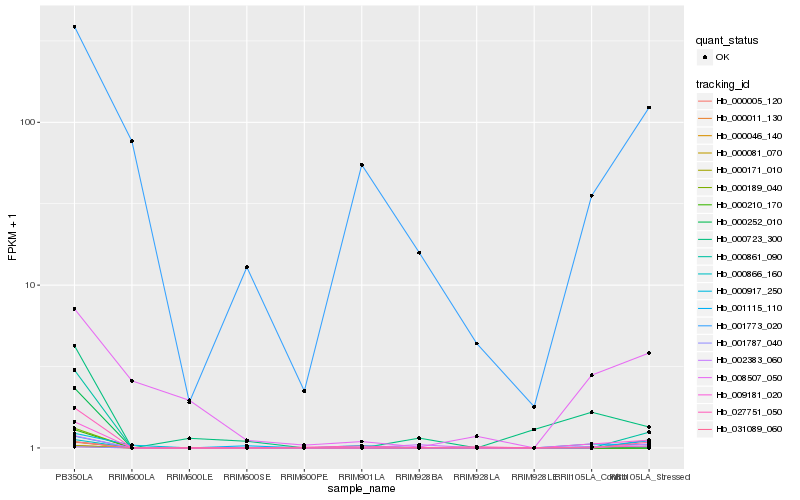
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_009181_020 |
0.0 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_000861_090 |
0.2459951011 |
- |
- |
- |
| 3 |
Hb_001115_110 |
0.2596770689 |
transcription factor |
TF Family: NAC |
NAC transcription factor 057 [Jatropha curcas] |
| 4 |
Hb_000723_300 |
0.3051606906 |
- |
- |
- |
| 5 |
Hb_027751_050 |
0.3299473128 |
- |
- |
hypothetical protein JCGZ_05887 [Jatropha curcas] |
| 6 |
Hb_031089_060 |
0.3304954504 |
- |
- |
- |
| 7 |
Hb_000866_160 |
0.3316153185 |
- |
- |
PREDICTED: elicitor-responsive protein 3 [Populus euphratica] |
| 8 |
Hb_001773_020 |
0.3591867662 |
- |
- |
hypothetical protein POPTR_0016s00980g [Populus trichocarpa] |
| 9 |
Hb_002383_060 |
0.360266611 |
- |
- |
PREDICTED: uncharacterized protein LOC102596491 [Solanum tuberosum] |
| 10 |
Hb_008507_050 |
0.3663123408 |
- |
- |
PREDICTED: S-adenosylmethionine decarboxylase proenzyme-like [Jatropha curcas] |
| 11 |
Hb_001787_040 |
0.3688399917 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_000917_250 |
0.3877095177 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Populus euphratica] |
| 13 |
Hb_000005_120 |
0.3955414362 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1 [Jatropha curcas] |
| 14 |
Hb_000011_130 |
0.3955414362 |
- |
- |
lipase [Ricinus communis] |
| 15 |
Hb_000046_140 |
0.3955414362 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000081_070 |
0.3955414362 |
- |
- |
- |
| 17 |
Hb_000171_010 |
0.3955414362 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 18 |
Hb_000189_040 |
0.3955414362 |
- |
- |
hypothetical protein [Trichoderma aureoviride] |
| 19 |
Hb_000210_170 |
0.3955414362 |
- |
- |
- |
| 20 |
Hb_000252_010 |
0.3955414362 |
- |
- |
- |