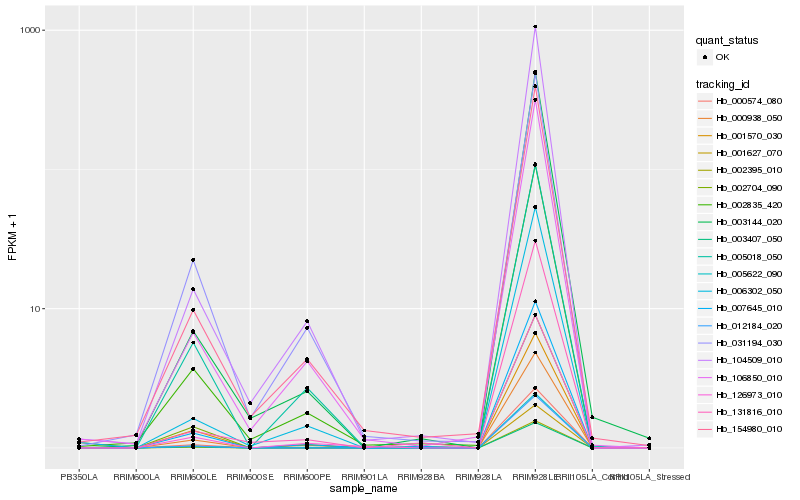
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007645_010 |
0.0 |
- |
- |
NdhE, partial (chloroplast) [Podocalyx loranthoides] |
| 2 |
Hb_002835_420 |
0.0752754553 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 3 |
Hb_106850_010 |
0.0804882114 |
- |
- |
photosystem II protein D2 [Lactuca sativa] |
| 4 |
Hb_000938_050 |
0.0807916114 |
- |
- |
hypothetical protein JCGZ_09640 [Jatropha curcas] |
| 5 |
Hb_154980_010 |
0.081433483 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 6 |
Hb_006302_050 |
0.08561098 |
- |
- |
PREDICTED: uncharacterized protein LOC104804969, partial [Tarenaya hassleriana] |
| 7 |
Hb_104509_010 |
0.0859740673 |
- |
- |
photosystem II CP43 chlorophyll apoprotein (chloroplast) [Ficus sp. Moore 315] |
| 8 |
Hb_126973_010 |
0.0869344048 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g34110 [Vitis vinifera] |
| 9 |
Hb_001627_070 |
0.090760152 |
transcription factor |
TF Family: MIKC |
PREDICTED: developmental protein SEPALLATA 1 [Nelumbo nucifera] |
| 10 |
Hb_002704_090 |
0.0908642942 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 11 |
Hb_031194_030 |
0.0912754349 |
- |
- |
ribulose 1,5-bisphosphate carboxylase/oxygenase [Stackhousia minima] |
| 12 |
Hb_000574_080 |
0.0914384313 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 13 |
Hb_012184_020 |
0.0914657119 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001570_030 |
0.0916143218 |
- |
- |
hypothetical protein JCGZ_13485 [Jatropha curcas] |
| 15 |
Hb_002395_010 |
0.0921359241 |
- |
- |
- |
| 16 |
Hb_131816_010 |
0.0922595296 |
- |
- |
PREDICTED: subtilisin-like protease SBT1.7 [Jatropha curcas] |
| 17 |
Hb_005622_090 |
0.0925522338 |
- |
- |
hypothetical protein CICLE_v10027074mg [Citrus clementina] |
| 18 |
Hb_005018_050 |
0.0932430983 |
- |
- |
PREDICTED: licodione synthase-like [Jatropha curcas] |
| 19 |
Hb_003144_020 |
0.0938662293 |
- |
- |
(R)-limonene synthase, putative [Ricinus communis] |
| 20 |
Hb_003407_050 |
0.0955060487 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |