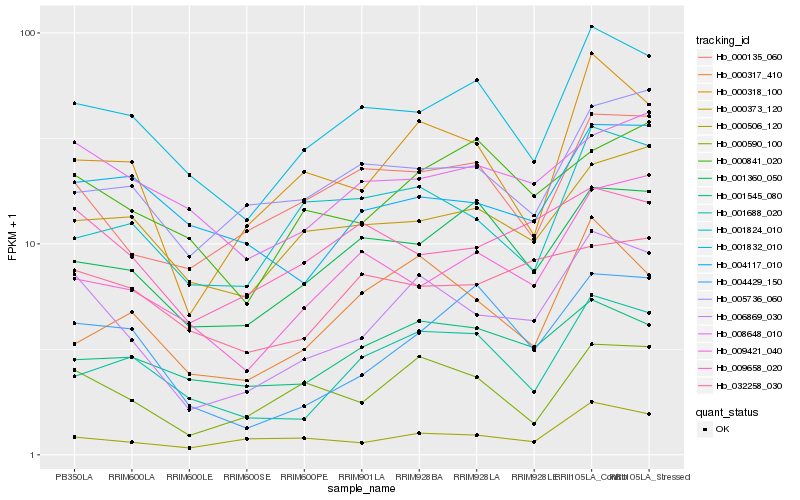
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006869_030 |
0.0 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 2 |
Hb_000318_100 |
0.13734126 |
transcription factor |
TF Family: G2-like |
PREDICTED: probable transcription factor KAN3 [Jatropha curcas] |
| 3 |
Hb_000590_100 |
0.1409745638 |
- |
- |
- |
| 4 |
Hb_001832_010 |
0.1506523676 |
- |
- |
PREDICTED: uncharacterized protein LOC105637890 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000506_120 |
0.1568662005 |
- |
- |
hypothetical protein AALP_AA6G154200 [Arabis alpina] |
| 6 |
Hb_000135_060 |
0.1630819441 |
transcription factor |
TF Family: MED6 |
PREDICTED: mediator of RNA polymerase II transcription subunit 6 [Jatropha curcas] |
| 7 |
Hb_004429_150 |
0.1636469764 |
- |
- |
- |
| 8 |
Hb_000373_120 |
0.1655028441 |
transcription factor |
TF Family: Trihelix |
PREDICTED: uncharacterized protein LOC105629120 [Jatropha curcas] |
| 9 |
Hb_001688_020 |
0.1670787095 |
- |
- |
PREDICTED: uncharacterized protein LOC103438026 [Malus domestica] |
| 10 |
Hb_008648_010 |
0.1700091017 |
- |
- |
PREDICTED: pre-rRNA-processing protein ESF2 [Jatropha curcas] |
| 11 |
Hb_009658_020 |
0.1703157513 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15 [Jatropha curcas] |
| 12 |
Hb_001360_050 |
0.1703847495 |
- |
- |
PREDICTED: probable helicase MAGATAMA 3 isoform X1 [Jatropha curcas] |
| 13 |
Hb_004117_010 |
0.1725274386 |
- |
- |
60S ribosomal protein L10, mitochondrial, putative [Ricinus communis] |
| 14 |
Hb_000317_410 |
0.1733116515 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C5-like [Jatropha curcas] |
| 15 |
Hb_001545_080 |
0.1750814872 |
- |
- |
PREDICTED: tRNA pseudouridine(38/39) synthase isoform X3 [Jatropha curcas] |
| 16 |
Hb_009421_040 |
0.1754224339 |
- |
- |
PREDICTED: eukaryotic initiation factor 4A [Jatropha curcas] |
| 17 |
Hb_000841_020 |
0.1755070312 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 10-B-like [Populus euphratica] |
| 18 |
Hb_001824_010 |
0.1759197668 |
- |
- |
glutathione-s-transferase theta, gst, putative [Ricinus communis] |
| 19 |
Hb_005736_060 |
0.1772078499 |
- |
- |
PREDICTED: ribosome maturation protein SBDS [Jatropha curcas] |
| 20 |
Hb_032258_030 |
0.1773864218 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |