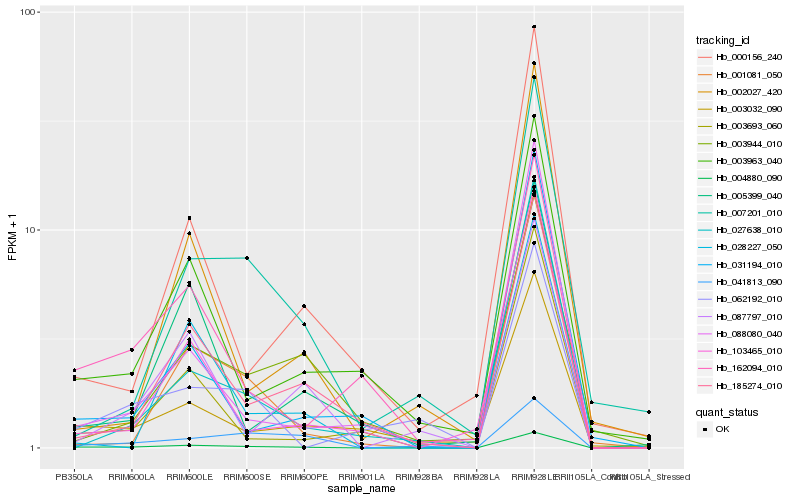
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004880_090 |
0.0 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 2 |
Hb_185274_010 |
0.1771631506 |
- |
- |
hypothetical protein ARALYDRAFT_330174 [Arabidopsis lyrata subsp. lyrata] |
| 3 |
Hb_003032_090 |
0.1856749055 |
- |
- |
hypothetical protein POPTR_0018s06450g [Populus trichocarpa] |
| 4 |
Hb_162094_010 |
0.1861455287 |
- |
- |
RNA polymerase alpha subunit [Hevea brasiliensis] |
| 5 |
Hb_001081_050 |
0.1891458292 |
- |
- |
hypothetical protein POPTR_0001s07860g [Populus trichocarpa] |
| 6 |
Hb_003963_040 |
0.1922819537 |
- |
- |
ATP-dependent Clp protease proteolytic subunit [Hevea brasiliensis] |
| 7 |
Hb_003693_060 |
0.1957004749 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit 6 [Medicago truncatula] |
| 8 |
Hb_103465_010 |
0.2049083644 |
- |
- |
RNA polymerase beta subunit [Hevea brasiliensis] |
| 9 |
Hb_007201_010 |
0.2055522304 |
- |
- |
PREDICTED: uncharacterized protein LOC105631226 isoform X2 [Jatropha curcas] |
| 10 |
Hb_027638_010 |
0.2056953866 |
- |
- |
Protein KES1, putative [Ricinus communis] |
| 11 |
Hb_028227_050 |
0.2075008168 |
- |
- |
PREDICTED: glucuronoxylan 4-O-methyltransferase 1 [Jatropha curcas] |
| 12 |
Hb_000156_240 |
0.2077341716 |
- |
- |
hypothetical protein MPER_07741 [Moniliophthora perniciosa FA553] |
| 13 |
Hb_087797_010 |
0.2090219542 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_088080_040 |
0.211461445 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit 2 A [Medicago truncatula] |
| 15 |
Hb_003944_010 |
0.2123719461 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 16 |
Hb_031194_010 |
0.2126952037 |
- |
- |
NADH-plastoquinone oxidoreductase subunit 2 (chloroplast) [Arachis hypogaea] |
| 17 |
Hb_062192_010 |
0.2142140192 |
- |
- |
- |
| 18 |
Hb_005399_040 |
0.215783588 |
- |
- |
RNA polymerase beta subunit [Hevea brasiliensis] |
| 19 |
Hb_002027_420 |
0.2206826402 |
- |
- |
hypothetical protein POPTR_0020s00240g [Populus trichocarpa] |
| 20 |
Hb_041813_090 |
0.2212287779 |
- |
- |
hypothetical protein EUGRSUZ_J01097 [Eucalyptus grandis] |