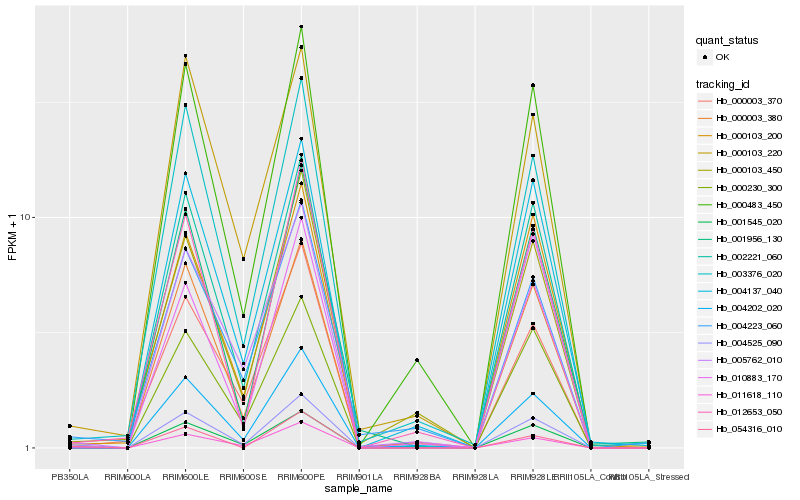
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004525_090 |
0.0 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 2 |
Hb_001545_020 |
0.0797435097 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X5 [Populus euphratica] |
| 3 |
Hb_002221_060 |
0.1094378008 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 4 |
Hb_000003_380 |
0.1105405262 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_010883_170 |
0.1128549357 |
- |
- |
PREDICTED: auxin transporter-like protein 3 [Jatropha curcas] |
| 6 |
Hb_000230_300 |
0.1170924446 |
- |
- |
PREDICTED: S-adenosylmethionine decarboxylase proenzyme 4 [Jatropha curcas] |
| 7 |
Hb_003376_020 |
0.1187580405 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 8 |
Hb_004202_020 |
0.1214952513 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_011618_110 |
0.121714238 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020-like [Citrus sinensis] |
| 10 |
Hb_012653_050 |
0.1220721461 |
- |
- |
hypothetical protein RCOM_0460020 [Ricinus communis] |
| 11 |
Hb_000103_200 |
0.1225144701 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 12 |
Hb_004137_040 |
0.1229510376 |
- |
- |
PREDICTED: serine carboxypeptidase-like 42 [Jatropha curcas] |
| 13 |
Hb_005762_010 |
0.1256078846 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic-like [Populus euphratica] |
| 14 |
Hb_000483_450 |
0.1256865971 |
- |
- |
PREDICTED: anthranilate N-benzoyltransferase protein 1 [Jatropha curcas] |
| 15 |
Hb_054316_010 |
0.1299022505 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |
| 16 |
Hb_000103_450 |
0.1302185107 |
- |
- |
PREDICTED: dentin sialophosphoprotein-like [Jatropha curcas] |
| 17 |
Hb_001956_130 |
0.1316092485 |
- |
- |
catalytic, putative [Ricinus communis] |
| 18 |
Hb_004223_060 |
0.1318781577 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |
| 19 |
Hb_000003_370 |
0.1337473133 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 3 [Jatropha curcas] |
| 20 |
Hb_000103_220 |
0.1346818661 |
- |
- |
PREDICTED: uncharacterized protein LOC105632036 [Jatropha curcas] |