| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
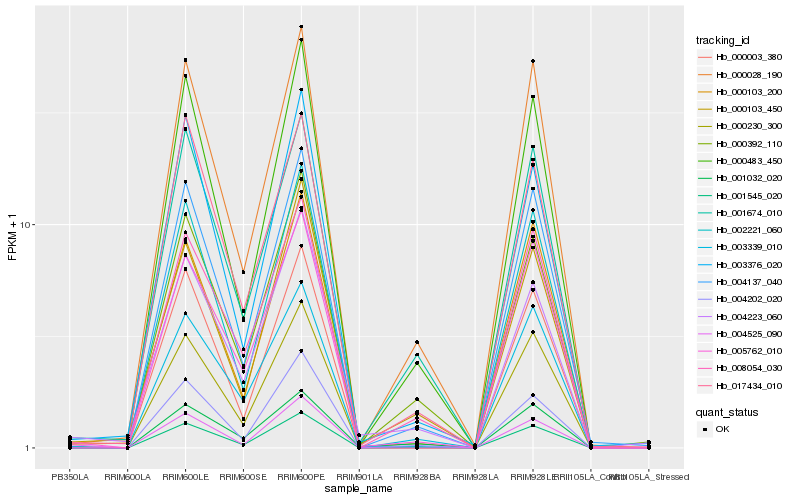
Hb_001545_020 |
0.0 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X5 [Populus euphratica] |
| 2 |
Hb_004137_040 |
0.0735144433 |
- |
- |
PREDICTED: serine carboxypeptidase-like 42 [Jatropha curcas] |
| 3 |
Hb_000028_190 |
0.0766806595 |
- |
- |
PREDICTED: probable inactive receptor kinase At1g48480 [Jatropha curcas] |
| 4 |
Hb_005762_010 |
0.0768962153 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic-like [Populus euphratica] |
| 5 |
Hb_004525_090 |
0.0797435097 |
transcription factor |
TF Family: C2C2-GATA |
GATA transcription factor, putative [Ricinus communis] |
| 6 |
Hb_000483_450 |
0.0800393992 |
- |
- |
PREDICTED: anthranilate N-benzoyltransferase protein 1 [Jatropha curcas] |
| 7 |
Hb_004202_020 |
0.0889576616 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001674_010 |
0.0894751187 |
- |
- |
PREDICTED: reticulon-like protein B21 isoform X1 [Populus euphratica] |
| 9 |
Hb_008054_030 |
0.0898858442 |
- |
- |
PREDICTED: VQ motif-containing protein 1-like [Jatropha curcas] |
| 10 |
Hb_002221_060 |
0.0915482059 |
transcription factor |
TF Family: GRAS |
Chitin-inducible gibberellin-responsive protein, putative [Ricinus communis] |
| 11 |
Hb_000103_200 |
0.0927742865 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 12 |
Hb_003339_010 |
0.0932508215 |
- |
- |
hypothetical protein JCGZ_04276 [Jatropha curcas] |
| 13 |
Hb_000230_300 |
0.0934025749 |
- |
- |
PREDICTED: S-adenosylmethionine decarboxylase proenzyme 4 [Jatropha curcas] |
| 14 |
Hb_000103_450 |
0.0936657931 |
- |
- |
PREDICTED: dentin sialophosphoprotein-like [Jatropha curcas] |
| 15 |
Hb_000392_110 |
0.0942409413 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 16 |
Hb_001032_020 |
0.0946096736 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 17 |
Hb_003376_020 |
0.0977469344 |
- |
- |
PREDICTED: interactor of constitutive active ROPs 2, chloroplastic isoform X1 [Jatropha curcas] |
| 18 |
Hb_000003_380 |
0.0977621207 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_017434_010 |
0.0988615497 |
- |
- |
PREDICTED: probable receptor protein kinase TMK1 [Jatropha curcas] |
| 20 |
Hb_004223_060 |
0.09934954 |
- |
- |
PREDICTED: WEB family protein At1g75720 [Jatropha curcas] |