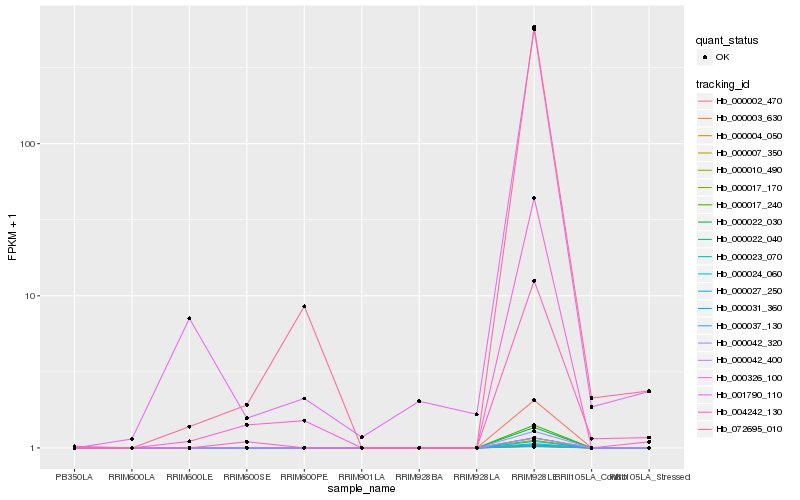
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004242_130 |
0.0 |
- |
- |
(-)-germacrene D synthase [Populus trichocarpa x Populus deltoides] |
| 2 |
Hb_072695_010 |
0.12266457 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 3 |
Hb_001790_110 |
0.1304617545 |
- |
- |
unknown [Populus trichocarpa] |
| 4 |
Hb_000326_100 |
0.131577061 |
transcription factor |
TF Family: MYB-related |
DnaJ homolog subfamily C member 2 [Morus notabilis] |
| 5 |
Hb_000002_470 |
0.1327647821 |
- |
- |
hypothetical protein JCGZ_11185 [Jatropha curcas] |
| 6 |
Hb_000003_630 |
0.1327647821 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 7 |
Hb_000004_050 |
0.1327647821 |
- |
- |
PREDICTED: uncharacterized protein LOC105766968 [Gossypium raimondii] |
| 8 |
Hb_000007_350 |
0.1327647821 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_000010_490 |
0.1327647821 |
- |
- |
hypothetical protein CICLE_v10027565mg, partial [Citrus clementina] |
| 10 |
Hb_000017_170 |
0.1327647821 |
transcription factor |
TF Family: FAR1 |
hypothetical protein MIMGU_mgv1a025104mg, partial [Erythranthe guttata] |
| 11 |
Hb_000017_240 |
0.1327647821 |
- |
- |
- |
| 12 |
Hb_000022_030 |
0.1327647821 |
- |
- |
Uncharacterized protein TCM_003129 [Theobroma cacao] |
| 13 |
Hb_000022_040 |
0.1327647821 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_000023_070 |
0.1327647821 |
- |
- |
hypothetical protein, partial [Cecembia lonarensis] |
| 15 |
Hb_000024_060 |
0.1327647821 |
- |
- |
PREDICTED: uncharacterized protein LOC105637052 [Jatropha curcas] |
| 16 |
Hb_000027_250 |
0.1327647821 |
transcription factor |
TF Family: FAR1 |
hypothetical protein MIMGU_mgv1a025104mg, partial [Erythranthe guttata] |
| 17 |
Hb_000031_360 |
0.1327647821 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_000037_130 |
0.1327647821 |
- |
- |
PREDICTED: uncharacterized protein LOC105775263 [Gossypium raimondii] |
| 19 |
Hb_000042_320 |
0.1327647821 |
- |
- |
unnamed protein product [Coffea canephora] |
| 20 |
Hb_000042_400 |
0.1327647821 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 5-like [Nicotiana tomentosiformis] |