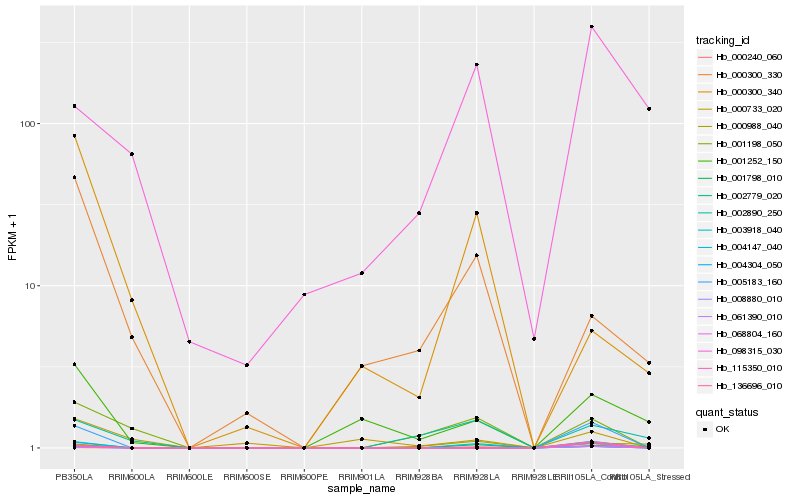
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004147_040 |
0.0 |
- |
- |
hypothetical protein JCGZ_15918 [Jatropha curcas] |
| 2 |
Hb_001798_010 |
0.3272720906 |
transcription factor |
TF Family: NAC |
NAC domain containing protein 2-like protein, partial [Helianthus annuus] |
| 3 |
Hb_000240_060 |
0.3438989977 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 4 |
Hb_001198_050 |
0.3447173203 |
- |
- |
hypothetical protein POPTR_0004s22990g [Populus trichocarpa] |
| 5 |
Hb_002890_250 |
0.3799617968 |
- |
- |
PREDICTED: uncharacterized protein LOC102611387 isoform X2 [Citrus sinensis] |
| 6 |
Hb_003918_040 |
0.3823423064 |
- |
- |
- |
| 7 |
Hb_005183_160 |
0.3830134631 |
- |
- |
- |
| 8 |
Hb_004304_050 |
0.383539376 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 9 |
Hb_001252_150 |
0.3962036797 |
- |
- |
- |
| 10 |
Hb_000988_040 |
0.4053607691 |
- |
- |
- |
| 11 |
Hb_098315_030 |
0.4128652727 |
- |
- |
PREDICTED: putative glutamine amidotransferase YLR126C [Jatropha curcas] |
| 12 |
Hb_008880_010 |
0.4158976713 |
- |
- |
PREDICTED: receptor-like serine/threonine-protein kinase SD1-8 isoform X2 [Nelumbo nucifera] |
| 13 |
Hb_061390_010 |
0.4159458273 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 14 |
Hb_000300_330 |
0.416176175 |
- |
- |
- |
| 15 |
Hb_115350_010 |
0.4165210502 |
- |
- |
PREDICTED: uncharacterized protein LOC105796032 [Gossypium raimondii] |
| 16 |
Hb_002779_020 |
0.4166900199 |
- |
- |
transcription factor, putative [Ricinus communis] |
| 17 |
Hb_136696_010 |
0.417004791 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 18 |
Hb_068804_160 |
0.4219199322 |
transcription factor |
TF Family: MIKC |
hypothetical protein JCGZ_12632 [Jatropha curcas] |
| 19 |
Hb_000733_020 |
0.4332643162 |
- |
- |
- |
| 20 |
Hb_000300_340 |
0.4362114625 |
transcription factor |
TF Family: SET |
unnamed protein product [Vitis vinifera] |