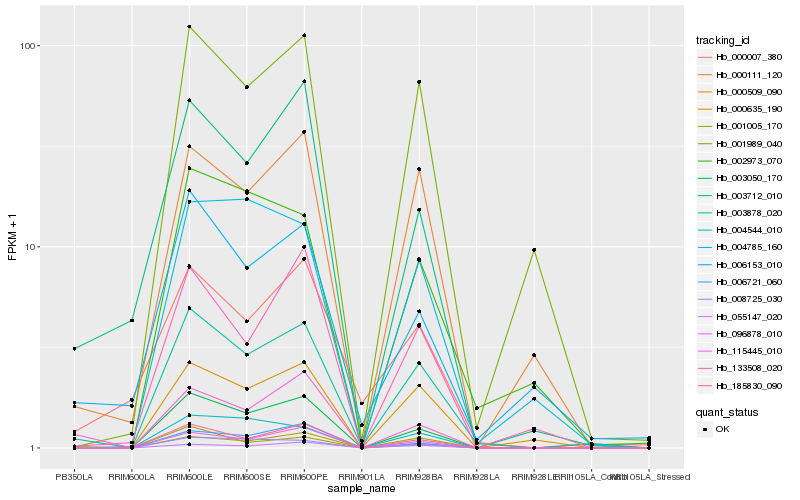
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003878_020 |
0.0 |
- |
- |
Epidermis-specific secreted glycoprotein EP1 precursor, putative [Ricinus communis] |
| 2 |
Hb_003050_170 |
0.1254205184 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_185830_090 |
0.1339330845 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001989_040 |
0.14628488 |
- |
- |
PREDICTED: neurofilament heavy polypeptide [Jatropha curcas] |
| 5 |
Hb_096878_010 |
0.1497732283 |
- |
- |
hypothetical protein POPTR_0112s002001g, partial [Populus trichocarpa] |
| 6 |
Hb_000509_090 |
0.151481059 |
- |
- |
Reticuline oxidase precursor, putative [Ricinus communis] |
| 7 |
Hb_000111_120 |
0.1558651065 |
- |
- |
Aspartic proteinase nepenthesin-1 precursor, putative [Ricinus communis] |
| 8 |
Hb_000635_190 |
0.1571744806 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103322127 [Prunus mume] |
| 9 |
Hb_001005_170 |
0.167054281 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase RPK2 [Jatropha curcas] |
| 10 |
Hb_008725_030 |
0.17097538 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003712_010 |
0.1710051731 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 12 |
Hb_004544_010 |
0.1737396158 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Jatropha curcas] |
| 13 |
Hb_006153_010 |
0.1748127783 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKU5 [Populus euphratica] |
| 14 |
Hb_133508_020 |
0.1753498878 |
- |
- |
PREDICTED: L-ascorbate oxidase homolog [Jatropha curcas] |
| 15 |
Hb_002973_070 |
0.1759207792 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 40 [Jatropha curcas] |
| 16 |
Hb_115445_010 |
0.1769594366 |
- |
- |
- |
| 17 |
Hb_000007_380 |
0.1820508892 |
- |
- |
Disease resistance family protein / LRR family protein, putative [Theobroma cacao] |
| 18 |
Hb_006721_060 |
0.183586715 |
- |
- |
RnaseH (mitochondrion) [Malus domestica] |
| 19 |
Hb_055147_020 |
0.1839357682 |
- |
- |
hypothetical protein EUGRSUZ_C03837 [Eucalyptus grandis] |
| 20 |
Hb_004785_160 |
0.1851314394 |
- |
- |
PREDICTED: U-box domain-containing protein 28-like [Jatropha curcas] |