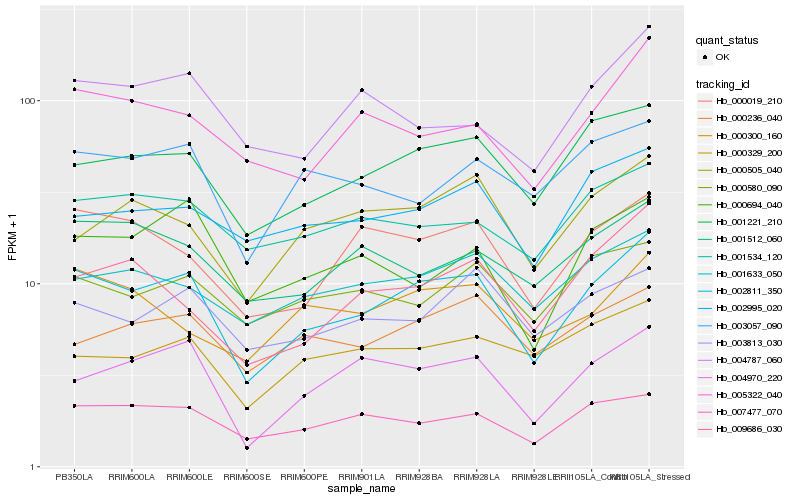
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002811_350 |
0.0 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 2 |
Hb_001221_210 |
0.1001580012 |
- |
- |
thiF family protein [Populus trichocarpa] |
| 3 |
Hb_000019_210 |
0.1111126585 |
transcription factor |
TF Family: GNAT |
PREDICTED: N-alpha-acetyltransferase MAK3 [Jatropha curcas] |
| 4 |
Hb_007477_070 |
0.1114251025 |
- |
- |
hypothetical protein JCGZ_17593 [Jatropha curcas] |
| 5 |
Hb_004970_220 |
0.1120747924 |
- |
- |
PREDICTED: uncharacterized protein LOC105633642 [Jatropha curcas] |
| 6 |
Hb_005322_040 |
0.1210733713 |
- |
- |
60S ribosomal protein L9, putative [Ricinus communis] |
| 7 |
Hb_001633_050 |
0.1216848063 |
- |
- |
PREDICTED: histone H2A.Z-specific chaperone CHZ1 [Jatropha curcas] |
| 8 |
Hb_003813_030 |
0.125138391 |
- |
- |
PREDICTED: mannose-6-phosphate isomerase 1 [Jatropha curcas] |
| 9 |
Hb_009686_030 |
0.1255360136 |
- |
- |
- |
| 10 |
Hb_002995_020 |
0.1260100113 |
transcription factor |
TF Family: Alfin-like |
PREDICTED: PHD finger protein ALFIN-LIKE 2 [Jatropha curcas] |
| 11 |
Hb_001534_120 |
0.1263610856 |
- |
- |
PREDICTED: vacuolar protein-sorting-associated protein 37 homolog 1 [Gossypium raimondii] |
| 12 |
Hb_003057_090 |
0.1270608258 |
- |
- |
ATP synthase D chain, mitochondrial, putative [Ricinus communis] |
| 13 |
Hb_000505_040 |
0.128130638 |
- |
- |
- |
| 14 |
Hb_000236_040 |
0.1281456501 |
- |
- |
PREDICTED: uncharacterized protein LOC105633150 isoform X1 [Jatropha curcas] |
| 15 |
Hb_001512_060 |
0.1282676616 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g35850, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000694_040 |
0.1297474396 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_000580_090 |
0.1299350314 |
- |
- |
PREDICTED: tRNA 2'-phosphotransferase 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000329_200 |
0.1306709679 |
- |
- |
PREDICTED: uncharacterized protein LOC105639316 [Jatropha curcas] |
| 19 |
Hb_004787_060 |
0.1307410747 |
- |
- |
PREDICTED: 40S ribosomal protein S9-2 [Jatropha curcas] |
| 20 |
Hb_000300_160 |
0.1309749753 |
transcription factor |
TF Family: C2C2-Dof |
PREDICTED: cyclic dof factor 3-like [Jatropha curcas] |