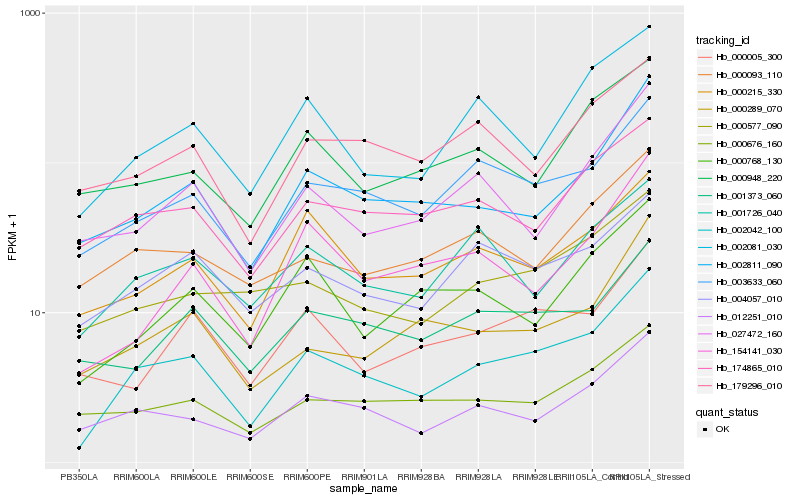
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002042_100 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647565 [Jatropha curcas] |
| 2 |
Hb_003633_060 |
0.1315322056 |
- |
- |
translationally controlled tumor protein [Hevea brasiliensis] |
| 3 |
Hb_012251_010 |
0.135420149 |
- |
- |
- |
| 4 |
Hb_002811_090 |
0.1356371651 |
- |
- |
40S ribosomal protein S24A [Hevea brasiliensis] |
| 5 |
Hb_002081_030 |
0.1441832461 |
- |
- |
unnamed protein product [Coffea canephora] |
| 6 |
Hb_000215_330 |
0.1549355131 |
- |
- |
PREDICTED: AP-4 complex subunit sigma [Jatropha curcas] |
| 7 |
Hb_174865_010 |
0.1552389668 |
- |
- |
transcription initiation factor iia (tfiia), gamma chain, putative [Ricinus communis] |
| 8 |
Hb_000005_300 |
0.1556533523 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_027472_160 |
0.1564191517 |
- |
- |
PREDICTED: proteasome subunit alpha type-2-A [Jatropha curcas] |
| 10 |
Hb_000289_070 |
0.1572067759 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000577_090 |
0.1584735187 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_154141_030 |
0.1584858113 |
- |
- |
PREDICTED: histone H2AX [Populus euphratica] |
| 13 |
Hb_004057_010 |
0.1585485528 |
- |
- |
PREDICTED: uncharacterized protein LOC105649313 [Jatropha curcas] |
| 14 |
Hb_000676_160 |
0.1585918359 |
- |
- |
folate carrier protein, putative [Ricinus communis] |
| 15 |
Hb_000768_130 |
0.1596052769 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 16 |
Hb_001726_040 |
0.1602555579 |
- |
- |
hypothetical protein CICLE_v10029230mg [Citrus clementina] |
| 17 |
Hb_000093_110 |
0.1605221516 |
- |
- |
PREDICTED: cytochrome b5 isoform A [Jatropha curcas] |
| 18 |
Hb_001373_060 |
0.1612585063 |
- |
- |
PREDICTED: structure-specific endonuclease subunit slx1 [Jatropha curcas] |
| 19 |
Hb_179296_010 |
0.1649406467 |
- |
- |
hypothetical protein B456_006G094200 [Gossypium raimondii] |
| 20 |
Hb_000948_220 |
0.1652634971 |
- |
- |
hypothetical protein CICLE_v10013189mg [Citrus clementina] |