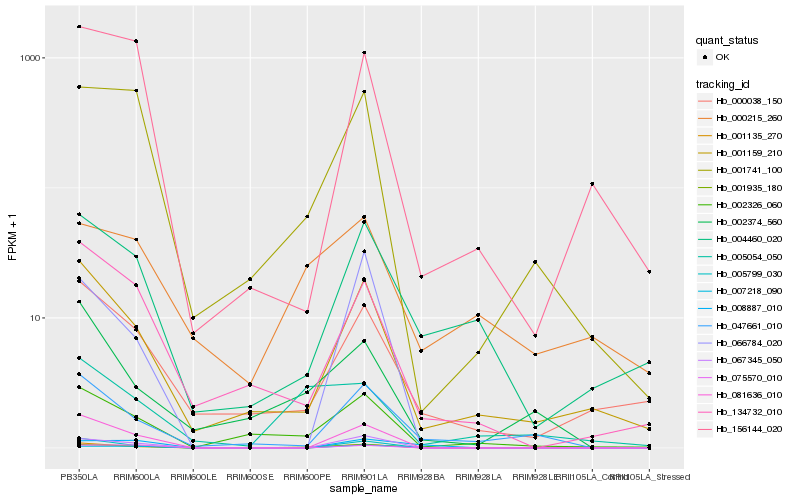
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001935_180 |
0.0 |
- |
- |
hypothetical protein VITISV_028082 [Vitis vinifera] |
| 2 |
Hb_001741_100 |
0.2427896854 |
rubber biosynthesis |
Gene Name: REF6 |
unnamed protein product [Vitis vinifera] |
| 3 |
Hb_007218_090 |
0.2439222993 |
- |
- |
PREDICTED: uncharacterized protein LOC104214925 [Nicotiana sylvestris] |
| 4 |
Hb_008887_010 |
0.2537560496 |
- |
- |
- |
| 5 |
Hb_002326_060 |
0.2541292666 |
- |
- |
PREDICTED: pollen-specific leucine-rich repeat extensin-like protein 3 [Populus euphratica] |
| 6 |
Hb_004460_020 |
0.2614254347 |
- |
- |
chaperone protein DNAj, putative [Ricinus communis] |
| 7 |
Hb_067345_050 |
0.2662246292 |
- |
- |
PREDICTED: uncharacterized protein LOC104107076 [Nicotiana tomentosiformis] |
| 8 |
Hb_001159_210 |
0.2709469963 |
- |
- |
PREDICTED: MATE efflux family protein DTX1 [Jatropha curcas] |
| 9 |
Hb_134732_010 |
0.2720621143 |
- |
- |
- |
| 10 |
Hb_000038_150 |
0.2753345513 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RNF181 homolog [Jatropha curcas] |
| 11 |
Hb_081636_010 |
0.2754440225 |
- |
- |
- |
| 12 |
Hb_001135_270 |
0.2802173365 |
- |
- |
- |
| 13 |
Hb_047661_010 |
0.2859777171 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 14 |
Hb_005799_030 |
0.2872507432 |
- |
- |
PREDICTED: uncharacterized protein LOC103926623 [Pyrus x bretschneideri] |
| 15 |
Hb_005054_050 |
0.2894550036 |
- |
- |
PREDICTED: uncharacterized protein At1g04910-like [Jatropha curcas] |
| 16 |
Hb_075570_010 |
0.2895654243 |
- |
- |
- |
| 17 |
Hb_000215_260 |
0.2924868151 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: leucine-rich repeat extensin-like protein 4 [Jatropha curcas] |
| 18 |
Hb_156144_020 |
0.2974972087 |
- |
- |
PREDICTED: uncharacterized protein At5g65660 [Vitis vinifera] |
| 19 |
Hb_066784_020 |
0.2975976923 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_002374_560 |
0.2990846955 |
- |
- |
PREDICTED: phosphatidylinositol/phosphatidylcholine transfer protein SFH11-like [Populus euphratica] |