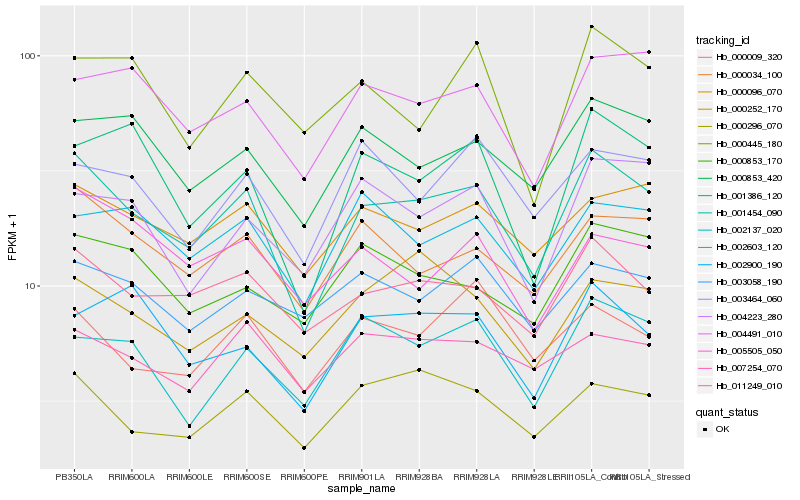
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001454_090 |
0.0 |
- |
- |
PREDICTED: alanyl-tRNA editing protein Aarsd1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_011249_010 |
0.0918154136 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 35, NatC auxiliary subunit isoform X1 [Jatropha curcas] |
| 3 |
Hb_000034_100 |
0.1000224572 |
- |
- |
dnajc14 protein, putative [Ricinus communis] |
| 4 |
Hb_000853_420 |
0.1023795802 |
- |
- |
PREDICTED: phosphoribosylformylglycinamidine cyclo-ligase, chloroplastic/mitochondrial [Jatropha curcas] |
| 5 |
Hb_000096_070 |
0.1053701352 |
- |
- |
PREDICTED: ruvB-like 2 [Jatropha curcas] |
| 6 |
Hb_001386_120 |
0.1066354981 |
- |
- |
PREDICTED: yrdC domain-containing protein, mitochondrial [Jatropha curcas] |
| 7 |
Hb_000296_070 |
0.1069633711 |
- |
- |
hypothetical protein B456_003G048700 [Gossypium raimondii] |
| 8 |
Hb_004491_010 |
0.1084804358 |
- |
- |
60S ribosomal protein L51, putative [Ricinus communis] |
| 9 |
Hb_002603_120 |
0.1153096861 |
- |
- |
PREDICTED: nuclear pore complex protein NUP88 [Jatropha curcas] |
| 10 |
Hb_002137_020 |
0.1158104953 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 11 |
Hb_000853_170 |
0.1160054461 |
- |
- |
PREDICTED: programmed cell death protein 2 isoform X3 [Populus euphratica] |
| 12 |
Hb_000009_320 |
0.117638855 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g66631 [Jatropha curcas] |
| 13 |
Hb_007254_070 |
0.1186804808 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase HERC2-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_002900_190 |
0.1189495726 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GTL1 [Jatropha curcas] |
| 15 |
Hb_003464_060 |
0.1192352465 |
- |
- |
PREDICTED: F-box protein PP2-A15 [Jatropha curcas] |
| 16 |
Hb_003058_190 |
0.1199760658 |
- |
- |
PREDICTED: uncharacterized protein LOC105628951 isoform X1 [Jatropha curcas] |
| 17 |
Hb_004223_280 |
0.1203187396 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger CCCH domain-containing protein 6 [Jatropha curcas] |
| 18 |
Hb_000445_180 |
0.1232596153 |
transcription factor |
TF Family: TRAF |
PREDICTED: BTB/POZ domain-containing protein At4g08455 [Jatropha curcas] |
| 19 |
Hb_000252_170 |
0.1236126911 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |
| 20 |
Hb_005505_050 |
0.1237420482 |
- |
- |
PREDICTED: uncharacterized protein LOC105639309 isoform X2 [Jatropha curcas] |