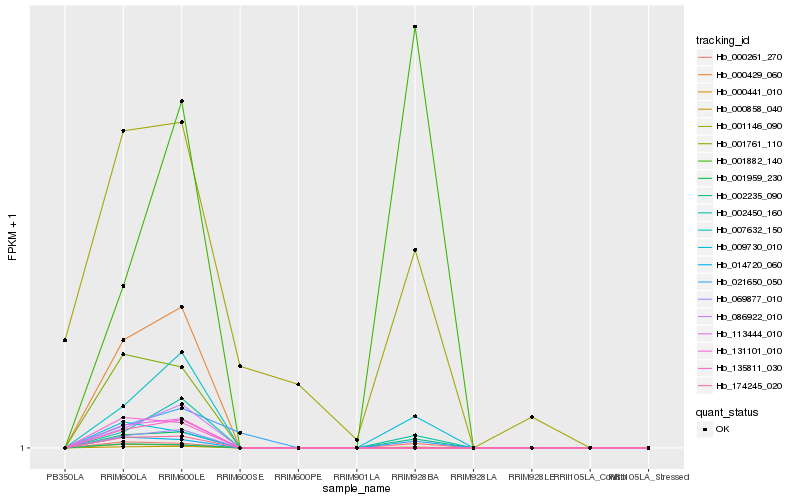
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000858_040 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 2 |
Hb_000261_270 |
0.1413993621 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 3 |
Hb_002450_160 |
0.1614789873 |
- |
- |
PREDICTED: protein 108 [Jatropha curcas] |
| 4 |
Hb_009730_010 |
0.2459515453 |
- |
- |
- |
| 5 |
Hb_001882_140 |
0.3020819589 |
- |
- |
- |
| 6 |
Hb_001146_090 |
0.3113858542 |
- |
- |
valacyclovir hydrolase, putative [Ricinus communis] |
| 7 |
Hb_002235_090 |
0.3117727814 |
- |
- |
UDP-glucuronosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_021650_050 |
0.3402748747 |
- |
- |
hypothetical protein RCOM_0852210 [Ricinus communis] |
| 9 |
Hb_069877_010 |
0.3477164709 |
- |
- |
chaperone protein DNAj, putative [Ricinus communis] |
| 10 |
Hb_000429_060 |
0.3477638949 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_135811_030 |
0.3478325264 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 12 |
Hb_113444_010 |
0.348620906 |
- |
- |
- |
| 13 |
Hb_001959_230 |
0.3493716098 |
- |
- |
PREDICTED: uncharacterized protein LOC105638436 [Jatropha curcas] |
| 14 |
Hb_174245_020 |
0.3518031105 |
- |
- |
- |
| 15 |
Hb_086922_010 |
0.3587283353 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 16 |
Hb_007632_150 |
0.363038273 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001761_110 |
0.3652481185 |
- |
- |
Contains similarity to Cf-2.2 gene gb |
| 18 |
Hb_131101_010 |
0.3665191815 |
- |
- |
- |
| 19 |
Hb_014720_060 |
0.3712789619 |
- |
- |
hypothetical protein JCGZ_11469 [Jatropha curcas] |
| 20 |
Hb_000441_010 |
0.3715162051 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |