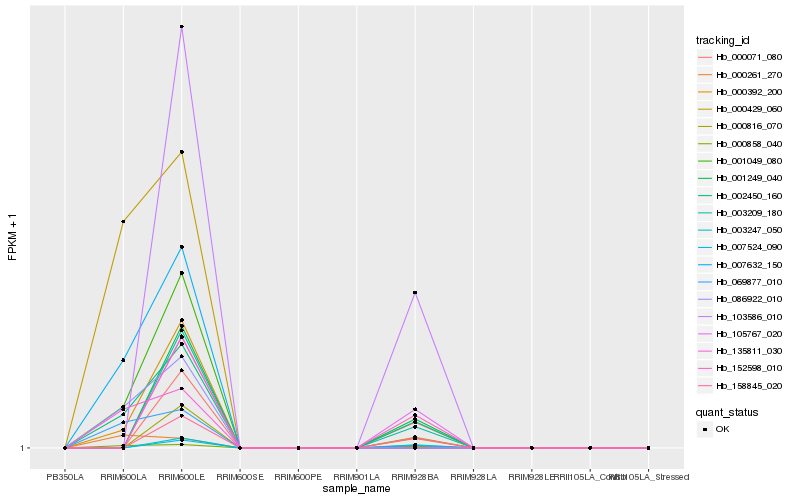
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002450_160 |
0.0 |
- |
- |
PREDICTED: protein 108 [Jatropha curcas] |
| 2 |
Hb_000858_040 |
0.1614789873 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 3 |
Hb_007632_150 |
0.2878387497 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001049_080 |
0.2907491079 |
- |
- |
- |
| 5 |
Hb_086922_010 |
0.2908685855 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 6 |
Hb_000261_270 |
0.2999890041 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 7 |
Hb_000392_200 |
0.3126842102 |
- |
- |
PREDICTED: cytochrome P450 98A2-like isoform X2 [Phoenix dactylifera] |
| 8 |
Hb_135811_030 |
0.3167730158 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 9 |
Hb_069877_010 |
0.3187357376 |
- |
- |
chaperone protein DNAj, putative [Ricinus communis] |
| 10 |
Hb_000816_070 |
0.3247954263 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001249_040 |
0.3250439149 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 12 |
Hb_152598_010 |
0.3256405301 |
- |
- |
PREDICTED: uncharacterized protein LOC104115096 [Nicotiana tomentosiformis] |
| 13 |
Hb_103586_010 |
0.3258179154 |
- |
- |
PREDICTED: serine/threonine-protein phosphatase 7 long form homolog [Glycine max] |
| 14 |
Hb_000429_060 |
0.326487627 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_158845_020 |
0.3276946785 |
transcription factor |
TF Family: TUB |
PREDICTED: tubby-like protein 8 [Jatropha curcas] |
| 16 |
Hb_000071_080 |
0.3282398981 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO5 [Jatropha curcas] |
| 17 |
Hb_003247_050 |
0.3283059009 |
- |
- |
- |
| 18 |
Hb_007524_090 |
0.3287306829 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 19 |
Hb_105767_020 |
0.3290634503 |
- |
- |
- |
| 20 |
Hb_003209_180 |
0.3290797375 |
- |
- |
Alpha-1,4 glucan phosphorylase L-2 isozyme [Morus notabilis] |