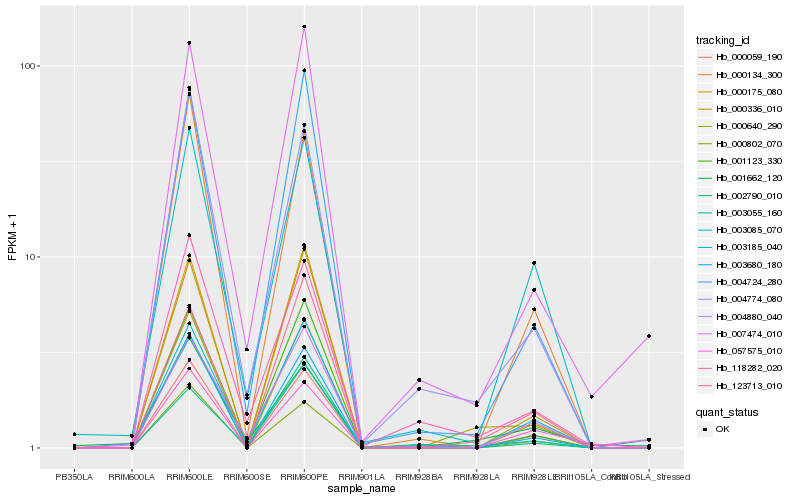
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000640_290 |
0.0 |
- |
- |
basic helix-loop-helix protein [Populus simonii x Populus nigra] |
| 2 |
Hb_000059_190 |
0.130539404 |
- |
- |
PREDICTED: uncharacterized protein LOC105121874 [Populus euphratica] |
| 3 |
Hb_123713_010 |
0.1424005331 |
- |
- |
o-methyltransferase, putative [Ricinus communis] |
| 4 |
Hb_003680_180 |
0.1436874127 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 5 |
Hb_004774_080 |
0.1440854394 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000802_070 |
0.1451962836 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: eugenol synthase 1-like [Jatropha curcas] |
| 7 |
Hb_000175_080 |
0.1467614447 |
- |
- |
hypothetical protein JCGZ_06123 [Jatropha curcas] |
| 8 |
Hb_000134_300 |
0.1495337199 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 9 |
Hb_003185_040 |
0.1511265487 |
- |
- |
PREDICTED: 3-ketoacyl-CoA synthase 11-like [Populus euphratica] |
| 10 |
Hb_000336_010 |
0.1511681443 |
transcription factor |
TF Family: C3H |
zinc finger family protein [Populus trichocarpa] |
| 11 |
Hb_004724_280 |
0.1545681514 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002790_010 |
0.1584009026 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 13 |
Hb_001123_330 |
0.1617494257 |
- |
- |
PREDICTED: serine carboxypeptidase-like 45 [Jatropha curcas] |
| 14 |
Hb_118282_020 |
0.1726147619 |
- |
- |
PREDICTED: uncharacterized protein LOC105631532 isoform X3 [Jatropha curcas] |
| 15 |
Hb_003085_070 |
0.1727365459 |
- |
- |
PREDICTED: BURP domain-containing protein 17-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_007474_010 |
0.1741396364 |
- |
- |
vacuolar invertase [Manihot esculenta] |
| 17 |
Hb_001662_120 |
0.1743590704 |
- |
- |
PREDICTED: ammonium transporter 3 member 1-like [Jatropha curcas] |
| 18 |
Hb_003055_160 |
0.1754071897 |
- |
- |
PREDICTED: S-linalool synthase isoform X1 [Jatropha curcas] |
| 19 |
Hb_057575_010 |
0.1757325084 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004880_040 |
0.1800963252 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g27290 isoform X1 [Jatropha curcas] |