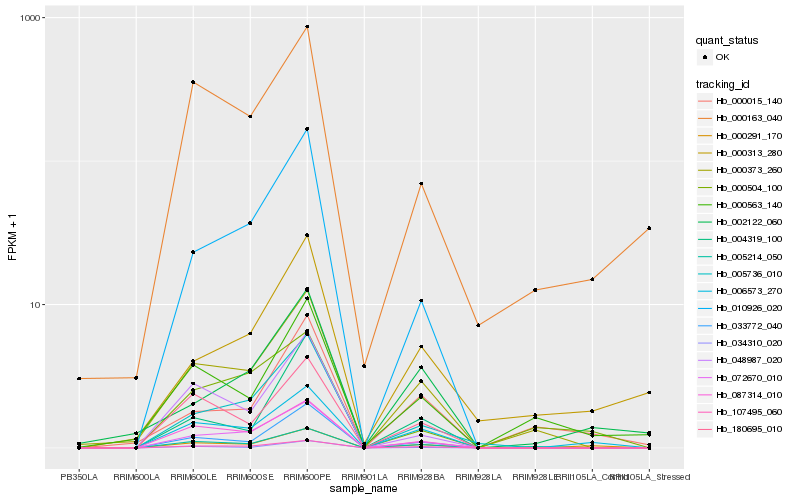
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000291_170 |
0.0 |
- |
- |
PREDICTED: inactive rhomboid protein 1 isoform X1 [Jatropha curcas] |
| 2 |
Hb_006573_270 |
0.0794175145 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_107495_060 |
0.1785882677 |
- |
- |
Cell division protein kinase, putative [Ricinus communis] |
| 4 |
Hb_033772_040 |
0.1915285096 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_072670_010 |
0.1970659677 |
- |
- |
Calcium-activated outward-rectifying potassium channel, putative [Ricinus communis] |
| 6 |
Hb_180695_010 |
0.2011649658 |
- |
- |
hypothetical protein CICLE_v10013449mg [Citrus clementina] |
| 7 |
Hb_087314_010 |
0.2013130376 |
- |
- |
- |
| 8 |
Hb_000015_140 |
0.2040878745 |
- |
- |
PREDICTED: CASP-like protein 4C1 [Vitis vinifera] |
| 9 |
Hb_000373_260 |
0.2080264364 |
- |
- |
hypothetical protein POPTR_0002s06820g [Populus trichocarpa] |
| 10 |
Hb_002122_060 |
0.210944976 |
- |
- |
PREDICTED: putative disease resistance protein RGA4 [Beta vulgaris subsp. vulgaris] |
| 11 |
Hb_034310_020 |
0.2110154731 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK3 [Jatropha curcas] |
| 12 |
Hb_005736_010 |
0.2132240135 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_010926_020 |
0.2151082179 |
- |
- |
PREDICTED: zinc-metallopeptidase, peroxisomal-like [Camelina sativa] |
| 14 |
Hb_000563_140 |
0.2153531778 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 15 |
Hb_000163_040 |
0.2206913563 |
- |
- |
PREDICTED: indole-3-acetic acid-induced protein ARG2-like [Jatropha curcas] |
| 16 |
Hb_004319_100 |
0.2209407752 |
- |
- |
PREDICTED: gibberellin-regulated protein 14 isoform X3 [Jatropha curcas] |
| 17 |
Hb_005214_050 |
0.2212856941 |
- |
- |
PREDICTED: leucine-rich repeat receptor protein kinase MSP1-like [Jatropha curcas] |
| 18 |
Hb_048987_020 |
0.2223530798 |
- |
- |
hypothetical protein JCGZ_27055 [Jatropha curcas] |
| 19 |
Hb_000313_280 |
0.2252992346 |
- |
- |
hypothetical protein PRUPE_ppa014856mg, partial [Prunus persica] |
| 20 |
Hb_000504_100 |
0.2291802865 |
- |
- |
conserved hypothetical protein [Ricinus communis] |