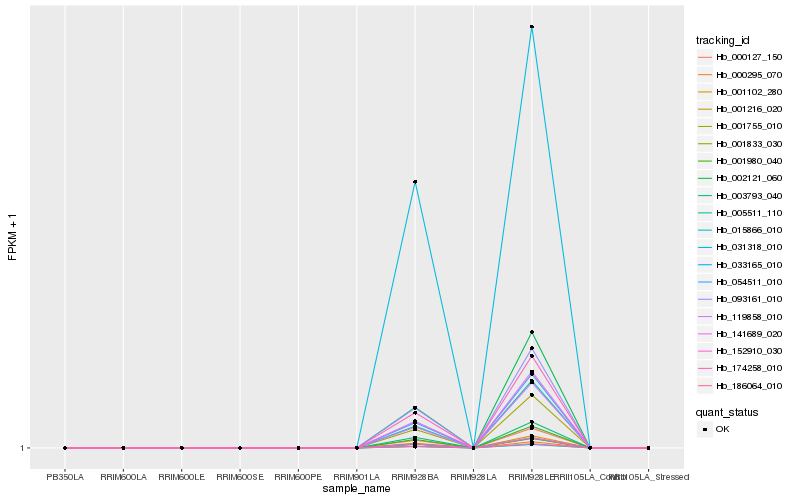
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_174258_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631548 [Jatropha curcas] |
| 2 |
Hb_001755_010 |
0.006310824 |
- |
- |
PREDICTED: uncharacterized protein LOC105767245 [Gossypium raimondii] |
| 3 |
Hb_093161_010 |
0.0064053937 |
- |
- |
- |
| 4 |
Hb_141689_020 |
0.0140622015 |
- |
- |
- |
| 5 |
Hb_054511_010 |
0.0165820086 |
- |
- |
Uncharacterized protein TCM_004340 [Theobroma cacao] |
| 6 |
Hb_033165_010 |
0.0214238999 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_186064_010 |
0.0220604472 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 8 |
Hb_005511_110 |
0.024509541 |
- |
- |
- |
| 9 |
Hb_001980_040 |
0.0251509283 |
- |
- |
- |
| 10 |
Hb_002121_060 |
0.0258652819 |
- |
- |
- |
| 11 |
Hb_001216_020 |
0.0269831962 |
- |
- |
retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa Japonica Group] |
| 12 |
Hb_119858_010 |
0.0272226755 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 13 |
Hb_001833_030 |
0.0274024637 |
- |
- |
polyprotein [Oryza australiensis] |
| 14 |
Hb_015866_010 |
0.0275510655 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 15 |
Hb_000127_150 |
0.027659183 |
- |
- |
PREDICTED: cytochrome P450 85A1-like [Jatropha curcas] |
| 16 |
Hb_152910_030 |
0.0279097565 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89B1-like [Jatropha curcas] |
| 17 |
Hb_031318_010 |
0.0279122801 |
- |
- |
- |
| 18 |
Hb_001102_280 |
0.0297353602 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 19 |
Hb_003793_040 |
0.0310539227 |
- |
- |
hypothetical protein CARUB_v10021783mg [Capsella rubella] |
| 20 |
Hb_000295_070 |
0.0312690891 |
- |
- |
polyprotein [Oryza australiensis] |