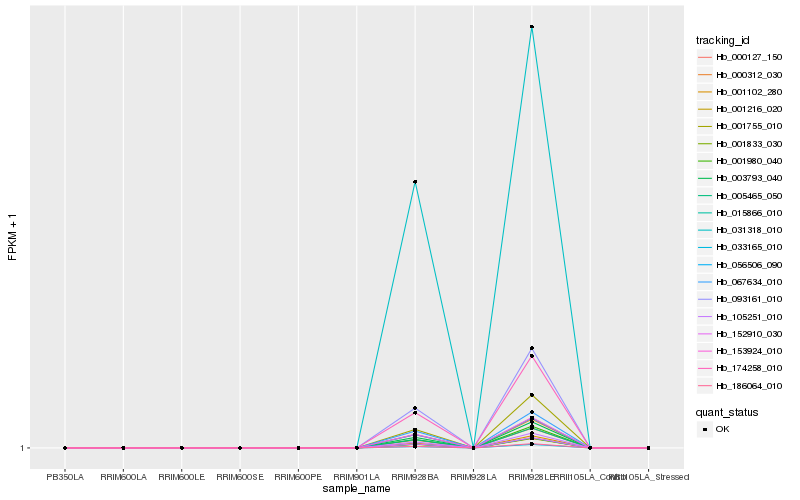
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_186064_010 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_033165_010 |
0.0006367123 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_001980_040 |
0.0030914165 |
- |
- |
- |
| 4 |
Hb_001216_020 |
0.0049243435 |
- |
- |
retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa Japonica Group] |
| 5 |
Hb_001833_030 |
0.0053437726 |
- |
- |
polyprotein [Oryza australiensis] |
| 6 |
Hb_015866_010 |
0.0054924327 |
- |
- |
PREDICTED: uncharacterized protein LOC105638675 [Jatropha curcas] |
| 7 |
Hb_000127_150 |
0.0056005929 |
- |
- |
PREDICTED: cytochrome P450 85A1-like [Jatropha curcas] |
| 8 |
Hb_152910_030 |
0.0058512665 |
- |
- |
PREDICTED: UDP-glycosyltransferase 89B1-like [Jatropha curcas] |
| 9 |
Hb_031318_010 |
0.0058537911 |
- |
- |
- |
| 10 |
Hb_001102_280 |
0.0076776423 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 11 |
Hb_003793_040 |
0.0089968095 |
- |
- |
hypothetical protein CARUB_v10021783mg [Capsella rubella] |
| 12 |
Hb_105251_010 |
0.0104518451 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_093161_010 |
0.015656292 |
- |
- |
- |
| 14 |
Hb_005465_050 |
0.0159079951 |
- |
- |
PREDICTED: uncharacterized protein LOC105635964 [Jatropha curcas] |
| 15 |
Hb_174258_010 |
0.0220604472 |
- |
- |
PREDICTED: uncharacterized protein LOC105631548 [Jatropha curcas] |
| 16 |
Hb_056506_090 |
0.0275953381 |
- |
- |
hypothetical protein B456_002G103400 [Gossypium raimondii] |
| 17 |
Hb_153924_010 |
0.0283466553 |
- |
- |
hypothetical protein JCGZ_22941 [Jatropha curcas] |
| 18 |
Hb_001755_010 |
0.0283690202 |
- |
- |
PREDICTED: uncharacterized protein LOC105767245 [Gossypium raimondii] |
| 19 |
Hb_000312_030 |
0.0352885187 |
- |
- |
PREDICTED: uncharacterized protein LOC104888072 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_067634_010 |
0.0355634508 |
- |
- |
hypothetical protein CICLE_v10006890mg, partial [Citrus clementina] |