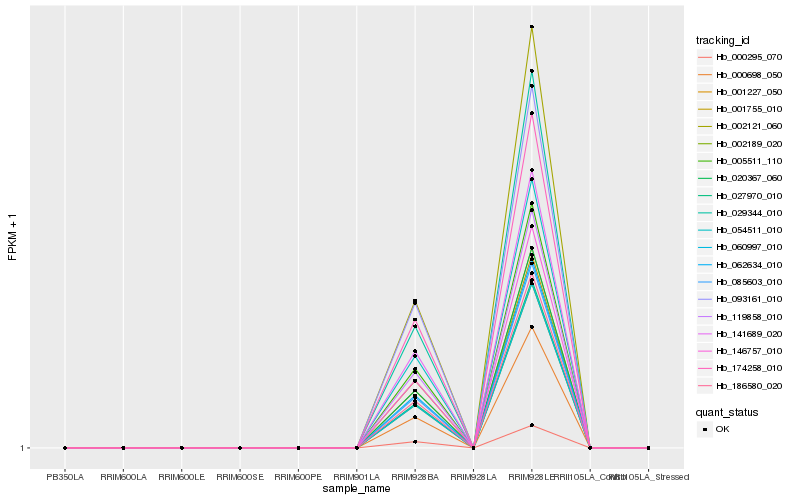
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002121_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_005511_110 |
0.0013562818 |
- |
- |
- |
| 3 |
Hb_119858_010 |
0.0013579978 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 4 |
Hb_000295_070 |
0.0054065874 |
- |
- |
polyprotein [Oryza australiensis] |
| 5 |
Hb_146757_010 |
0.0080001387 |
- |
- |
- |
| 6 |
Hb_054511_010 |
0.0092857483 |
- |
- |
Uncharacterized protein TCM_004340 [Theobroma cacao] |
| 7 |
Hb_029344_010 |
0.0116901472 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 8 |
Hb_141689_020 |
0.0118057387 |
- |
- |
- |
| 9 |
Hb_001227_050 |
0.017072861 |
- |
- |
- |
| 10 |
Hb_020367_060 |
0.017072861 |
- |
- |
hypothetical protein POPTR_0005s15010g [Populus trichocarpa] |
| 11 |
Hb_001755_010 |
0.0195564108 |
- |
- |
PREDICTED: uncharacterized protein LOC105767245 [Gossypium raimondii] |
| 12 |
Hb_062634_010 |
0.0216163927 |
- |
- |
PREDICTED: uncharacterized protein LOC105636643 [Jatropha curcas] |
| 13 |
Hb_085603_010 |
0.0232411993 |
- |
- |
PREDICTED: uncharacterized protein LOC101296995 [Fragaria vesca subsp. vesca] |
| 14 |
Hb_174258_010 |
0.0258652819 |
- |
- |
PREDICTED: uncharacterized protein LOC105631548 [Jatropha curcas] |
| 15 |
Hb_186580_020 |
0.0269016216 |
- |
- |
PREDICTED: uncharacterized protein LOC103426965 [Malus domestica] |
| 16 |
Hb_027970_010 |
0.0294777606 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 17 |
Hb_002189_020 |
0.0306656812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_060997_010 |
0.0306656812 |
- |
- |
PREDICTED: uncharacterized protein LOC105789525 [Gossypium raimondii] |
| 19 |
Hb_000698_050 |
0.0316776412 |
- |
- |
- |
| 20 |
Hb_093161_010 |
0.0322674672 |
- |
- |
- |